
Attempt to predict early recurrence of prostate cancer following prostatectomy through machine learning
In the developed world, prostate cancer (PC) is the most common malignancy in men and the second major cause of cancer deaths (1). The disease progresses from high grade prostatic intra-epithelial neoplasia (HGPIN) to carcinoma. Primary PCs can be managed with a variety of options including watchful waiting, radical prostatectomy (RP), and radiation. The choice of different management plans depends on disease severity, patient age and preference. PCs are graded with Gleason score (GS) and GS-based World Health Organization (WHO) PC grading system (WHO grade group 1–5) or International Society of Urological Pathology (ISUP) grade (2-4). The disease evolves with a high degree of disparity. While GS6/WHO grade group 1 tumors are generally indolent, higher grade PCs are at risk of progression. Approximately 30% of tumors will relapse following RP; recurrent PCs are commonly detected by a rise in serum prostate-specific antigen (PSA), a process that is characterized as biochemical recurrence (BCR) (5). BCR is a major progression of PC (6): approximately 40% of PCs with BCR will progress to metastatic disease, which is mainly treated with androgen-deprivation therapy (ADT). This treatment is generally palliative, as progression to metastatic castration resistant prostate cancer (mCRPC) inevitably occurs (7). In the last decades, numerous agents have been developed to treat mCRPCs, such as taxane-based chemotherapy and androgen receptor (AR)-targeting therapy involving either abiraterone or enzalutamide (8,9). These therapies modestly extend patient’s overall survival (OS) for a few months before resistance develops (10). Under the current situation, one option to improve patient management is through intervention at the BCR stage, which will likely be more effective than treatment of metastatic PCs. However, this strategy will require effective prediction of BCR risk.
The importance of stratification of PCs with elevated risk of recurrence has been well recognized; there are 2,294 publications listed on PubMed on September 2, 2018 under the search term of “Prostate cancer, biochemical recurrence, and biomarkers”. This extensive research effort has yielded two commercially available multi-gene (mRNA) panels, Oncotype DX (Genomic Prostate Score/GPS) and Prolaris [cell cycle progression (CCP)]. The 17-gene Oncotype DX and the 31-gene Prolaris both improve the prediction of PCs at risk of recurrence at time of diagnosis (11-15) and after RP (16,17). Recently, a 15-gene signature (SigMuc1NW) had been reported that robustly predicts BCR following prostatectomy (18). Even with these developments, there remains a clear need to improve the current stratification of PCs with high risk of recurrence.
To meet this need, Wong et al. reported an attractive system to assess the risk of early BCR in a group patient treated with robot-assisted prostatectomy (n=338) (19). This was a single center-based investigation using clinical materials of 338 patients who have been treated by robot-assisted prostatectomy for local PC by a single surgeon during May 2012 to Dec 2015. A group of 19 clinical variables have been collected (Table 1). PC is in general a slowly progressive disease; PC evolves with a high level of disparity. BCR develops from several months to years after RP (18,20). With their cohort composed of patients with modest length of follow-up and relatively small size (Figure 1), the authors focused on the prediction of early BCR that was developed within one year after RP. In their cohort (n=338), 25 patients had BCR (Figure 1) (19). Wong et al. have randomly divided the cohort into a training and testing population in a 7:3 ratio (19), and trained the training set for classification of early BCR (Figure 1). Four statistical machine learning systems [K-nearest neighbors, random forest, logistic regression, and Cox proportional hazards (PH) regression] were used to model the contributions of the 19 clinical variables (Table 1) to early BCR. The resultant models were than analyzed using the testing population (Figure 1). The models produced by K-nearest neighbors, random forest, and logistic regression were quite robust in the discrimination of BCR with the respective area under curve (AUC) value of 0.903, 0.924, and 0.94 (Figure 1). In comparison, the Cox PH model stratified early BCR with the AUC value of 0.865 (Figure 1) (19).
Table 1
| Baseline clinical variables | Association with BCR |
|---|---|
| Age | No |
| Body mass index | No |
| American Society of Anesthesiologists physical status classification | No2 |
| D’Amico risk group | Yes** |
| PSA | No3 |
| Gleason grade (biopsy) | Yes** |
| Prostate size on ultrasonography (volume) | No |
| Nerve-sparing status | Yes* |
| Estimated blood loss during surgery | No |
| Operating time | No |
| Length of hospital stay | No |
| Gleason grade (surgical) | Yes** |
| Percent volume of tumor involvement | Yes** |
| Extracapsular extension | Yes** |
| Seminal vesicle invasion | Yes** |
| Margin status | Yes* |
| T stage | Yes** |
| Number of nodes | No |
| Nodal involvement | Yes** |
1, association with BCR was determined using univariate Cox PH regression; 2, P=0.064; 3, P=0.078; *, P<0.05; **, P<0.01. BCR, biochemical recurrence; PSA, prostate-specific antigen.
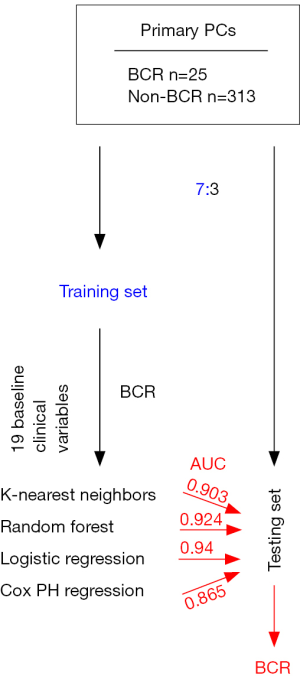
Machine learning has been rising as a powerful tool in classification and regression modeling of high dimensional variables in cancer recurrence and OS. For insistence, 150 clinical baseline variables have been modeled for prediction of OS in patients with mCRPC (21,22) and more than 600 differentially expressed genes have been selected to stratify BCR (18). The majority of these machine learning efforts were based on the Cox PH model. By formulating the response as with or without early BCR and ignoring the time-to-event component of BCR, Wong et al. used more flexible models K-nearest neighbors and random forest to model importance of the 19 baseline clinical variables (Table 1) in early BCR development (19). Both models require no hypothesis and no consideration of data distributions, are quite robust, and do not commonly produce overfitting models. In this regard, through simplification of PC recurrence by focusing on early BCR, both models and logistic regression can be robust. However, we should be cautious to conclude these models as superior to those of Cox PH-based; recurrence occurred at the first year is clearly not the same from those developed after 5 years. Even with recurrence within the first year, PCs that relapse within 6 months are likely different from those with recurrence progression at 12 months. Nonetheless, it can be envisaged that the kinetic issue can be minimized though more detailed division of recurrence timespan, for example 6, 12, 18 months and so on, following the growth in size and complexity of their patient population. Indeed, Wong et al. have proposed to expand their study with more patients and including additional clinical baseline factors. Additional patients can be recruited from other surgeons in their Institute. It will be more appealing if multiple centers can be involved in the future. The robustness of their model in the prediction of early BCR is calling this effort. Such efforts may lead to the generation of effective clinical systems to predict BCR. With today’s computing power and machine learning capacity, the days may not be too far for doctors to enter a set of baseline clinical variables at their terminals to come out with accurate prediction of PC recurrence. Clearly, the same principle can be applied to other clinical outcomes such as OS as well as other cancer types.
Despite the great potential discussed, this research is still at an early stage. One major limitation is the small sample size; the issue was compounded considering the random division of 25 recurrent tumors in a 7:3 ratio into a training and testing population. With the limited number of recurrent tumors in both the training and testing populations, the accuracy of the models will need to be confirmed using larger patient populations in the future.
It may shed light on the models with respect to their utility if more details of the model were provided. The models were built on 19 baseline clinical variables (Table 1), including those with well-established association with PC recurrence, such as Gleason scores, percentage of tumor involvement, extracapsular extension, seminal vesicle invasion, margin status, T-stage, and nodal involvement. Were all the 19 clinical variables essential or did those established clinical characteristics contribute more to the prediction? The feature (variable) importance derived from random forest modeling should provide an indication on this issue should this data be reported. Furthermore, lactate dehydrogenase (LDH), albumin, hemoglobin, alkaline phosphatase (ALP) (23) along with a set of clinical factors related to kidney function, haematology, and others (21,22) display predictive value toward OS in patients with mCRPC. Should these factors be relevant to the author’s models?
Progression to BCR is regulated by molecular networks; the complexity of these networks is clearly reflected by the number of publications (n=2,294) listed in PubMed (September 2, 2018) on this issue. The molecular alterations may also need to be included in the models reported in this study (19). A good starting point is to consider the genes reported in Oncotype DX (Genomic Prostate Score/GPS) (11-15), Prolaris (cell cycle progression/CCP) (16,17), and SigMuc1NW (18).
Acknowledgements
Funding: D.T. is supported by an Award from Teresa Cascioli Charitable Foundation Research Award in Women’s Health and grants from Canadian Cancer Society (grant #: 319412) and Cancer Research Society. Y.G. is supported by Studentship provided by Ontario Graduate Scholarships and Research Institute of St Joe’s Hamilton.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xiao Li (Department of Urologic Surgery, the Affiliated Cancer Hospital of Jiangsu Province of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2018.09.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Egevad L, Delahunt B, Srigley JR, et al. International Society of Urological Pathology (ISUP) grading of prostate cancer - An ISUP consensus on contemporary grading. APMIS 2016;124:433-5. [Crossref] [PubMed]
- Gordetsky J, Epstein J. Grading of prostatic adenocarcinoma: current state and prognostic implications. Diagn Pathol 2016;11:25. [Crossref] [PubMed]
- Epstein JI, Zelefsky MJ, Sjoberg DD, et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur Urol 2016;69:428-35. [Crossref] [PubMed]
- Zaorsky NG, Raj GV, Trabulsi EJ, et al. The dilemma of a rising prostate-specific antigen level after local therapy: what are our options? Semin Oncol 2013;40:322-36. [Crossref] [PubMed]
- Shipley WU, Seiferheld W, Lukka HR, et al. Radiation with or without Antiandrogen Therapy in Recurrent Prostate Cancer. N Engl J Med 2017;376:417-28. [Crossref] [PubMed]
- Semenas J, Allegrucci C, Boorjian SA, et al. Overcoming drug resistance and treating advanced prostate cancer. Curr Drug Targets 2012;13:1308-23. [Crossref] [PubMed]
- de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364:1995-2005. [Crossref] [PubMed]
- Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187-97. [Crossref] [PubMed]
- Ojo D, Lin X, Wong N, et al. Prostate Cancer Stem-like Cells Contribute to the Development of Castration-Resistant Prostate Cancer. Cancers (Basel) 2015;7:2290-308. [Crossref] [PubMed]
- Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics 2013;14:690. [Crossref] [PubMed]
- Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol 2014;66:550-60. [Crossref] [PubMed]
- Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245-55. [Crossref] [PubMed]
- Oderda M, Cozzi G, Daniele L, et al. Cell-cycle Progression-score Might Improve the Current Risk Assessment in Newly Diagnosed Prostate Cancer Patients. Urology 2017;102:73-8. [Crossref] [PubMed]
- Albala D, Kemeter MJ, Febbo PG, et al. Health Economic Impact and Prospective Clinical Utility of Oncotype DX(R) Genomic Prostate Score. Rev Urol 2016;18:123-32. [PubMed]
- Cullen J, Rosner IL, Brand TC, et al. A Biopsy-based 17-gene Genomic Prostate Score Predicts Recurrence After Radical Prostatectomy and Adverse Surgical Pathology in a Racially Diverse Population of Men with Clinically Low- and Intermediate-risk Prostate Cancer. Eur Urol 2015;68:123-31. [Crossref] [PubMed]
- Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol 2013;31:1428-34. [Crossref] [PubMed]
- Jiang Y, Mei W, Gu Y, et al. Construction of a set of novel and robust gene expression signatures predicting prostate cancer recurrence. Mol Oncol 2018;12:1559-78. [Crossref] [PubMed]
- Wong NC, Lam C, Patterson L, et al. Use of machine learning to predict early biochemical recurrence after robot-assisted prostatectomy. BJU Int 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Pound CR, Partin AW, Eisenberger MA, et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA 1999;281:1591-7. [Crossref] [PubMed]
- Guinney J, Wang T, Laajala TD, et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol 2017;18:132-42. [Crossref] [PubMed]
- Mei W, Kapoor A, Major P, et al. Progress towards accurate prediction of overall survival in men with metastatic castration-resistant prostate cancer. J Xiangya Med 2017;2:17. [Crossref]
- Halabi S, Lin CY, Kelly WK, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol 2014;32:671-7. [Crossref] [PubMed]
Cite this article as: Gu Y, Lin X, Kapoor A, Mei W, Tang D. Attempt to predict early recurrence of prostate cancer following prostatectomy through machine learning. AME Med J 2018;3:96.

