Hypoxia-inducible factors in cancer: an overview of major findings from meta-analyses
Introduction
Hypoxia, which refers to a low oxygen condition, is closely associated with the development and progression of cancer (1-3). Hypoxia-inducible factors (HIFs) are important proteins for the regulation of molecular response on hypoxia (4). HIFs consist of two subunits (i.e., α and β). The α subunit is expressed according to the oxygen conditions and determines the transcriptional activity of HIF. In details, the degradation of HIF-1α is enhanced and suppressed in the normoxic and hypoxic conditions, respectively; and high and low expression of HIF-1α increases and decreases the HIF activity, respectively. HIF-1α family contains 3 members (HIF-1α, HIF-2α, and HIF-3α) (5-8). By comparison, the β subunit is constitutively expressed in the nucleus.
Among the HIF-1α family members, HIF-1α is the most widely studied in human cancer (9,10). HIF-1α gene, which is located at the chromosome 14q21-24, consists of 15 exons and 14 introns, codes the cDNA of 3,919 bps, and produces the protein of 826 amino acids. HIF-1α can transactivate more than 70 target genes and is a master regulator of erythropoiesis, blood vessel formation, cell metabolism, and genetic stability. There are two major HIF-1α gene polymorphisms (C1772T and G1790A). Both of them are located at the exon 12 of the HIF-1α gene within the oxygen-dependent degradation domain. HIF-1α C1772T (rs11549465) mutation refers to an amino acid substitution from proline to serine at codon 582 (Pro582Ser or P582S). HIF-1α G1790A (rs11549467) mutation refers to an amino acid substitution from alanine to threonine at codon 588 (Ala588Thr or A588T).
To the best of our knowledge, numerous studies and meta-analyses have explored the role of HIF-1α gene polymorphism and protein expression in cancer. By comparison, less evidence has been accumulated regarding the role of HIF-2α and HIF-3α in cancer. In this paper, we have conducted an overview of meta-analyses to provide more comprehensive recognition of evidence regarding the role of HIFs in cancer.
Methods
Registration
Our study protocol was registered in PROSPERO database. The registration number was CRD42016037401.
Search strategy
We identified the relevant meta-analysis papers via the PubMed and EMBASE databases. We also manually identified the relevant meta-analysis papers. Search items were: “(hypoxia inducible factor) OR HIF” AND “(((cancer) OR tumor) OR neoplasm) OR carcinoma” AND “(meta analysis)”. The last search date was April 6, 2016.
Eligibility criteria
Only meta-analysis papers regarding the role of HIF in cancer were eligible for our study. Duplicates, comments or editorials, narrative reviews, original articles, and irrelevant meta-analysis papers were excluded. Publication language or date was not limited.
Data extraction
We primarily extracted the data from the eligible meta-analysis papers, as follows: first author, publication year, journal, country, databases which were employed for each meta-analysis, date when each meta-analysis was conducted, type of cancer, HIF gene polymorphism or protein expression, number of studies which were included in each meta-analysis, and results of each meta-analysis. If the statistical analyses were performed by using both fixed- and random-effects models, only the results by a random-effects model would be considered.
Evaluation of heterogeneity
If the results were heterogeneous among two or more meta-analyses, we would further identify the reliability according to the following criteria.
First, the number of eligible studies should be considered. A meta-analysis with a larger number of eligible studies would be more reliable.
Second, if the number of eligible studies was similar among them, the number of participants would be considered. A meta-analysis with a larger number of participants would be more reliable.
Third, if the eligible studies were completely overlapped among them, the methods of meta-analysis would be considered. A meta-analysis using a random-effect model would be more reliable.
Fourth, if the controversy or uncertainty remained according to the above-mentioned criteria, the original studies would be extracted and a meta-analysis might be updated. We might also contact with the authors or journal editors, if necessary.
Results
After excluding the irrelevant papers, a total of 55 meta-analysis papers were included in our study (Figure 1). Among them, 53 papers were written by Chinese researchers, 1 paper by UK researchers, and 1 paper by Bangladeshi researchers. The last search date for each meta-analysis ranged from 2009 to 2016. Results of meta-analyses were summarized according to the location of cancer.
Overall cancer
A total of 13 meta-analysis papers explored the role of HIF in overall cancer regardless of location of cancer (11-23) (Table S1). Among them, 5 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (11,13,15,18,22), 5 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (12,14,17,19,21), and 3 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20,23).
Risk
Nine papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of overall cancer (11,12,14,15,17-19,21,22). All of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of overall cancer (11,12,14,15,17-19,21,22).
Seven papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of overall cancer (11,15,16,18,20,22,23). Six of them demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of overall cancer (11,15,16,18,20,23). But another paper did not show any significant association between them (22). The meta-analyses by Liu P (16) and Zhou Y (23) had a larger number of included studies than those by Yang X (18), Ye Y (20), Anam MT (11), Zhao T (22), and Liu J (15) (26 and 26 versus 24, 21, 19, 12, and 6). Thus, we should support a significant association between HIF-1α rs11549467 (1790 G/A) polymorphism and the risk of overall cancer.
Clinicopathological features
One paper explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the clinicopathological features of overall cancer (13). It demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the lymph node metastasis and histological grade of overall cancer, but not the tumor size or stage (13).
One paper explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the clinicopathological features of overall cancer (13). It demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the lymph node metastasis and tumor size of overall cancer, but not the histological grade or tumor stage (13).
Head and neck cancer
A total of 4 meta-analysis papers explored the role of HIF in head and neck cancer (12,14,16,23) (Table S2). Among them, 2 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (12,14), and another 2 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,23).
Risk
Two papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of head and neck cancer (12,14). One of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of head and neck cancer (12). But another paper did not show any significant association between them (14). The meta-analysis by He P (12) had a larger number of included studies than that by Li Y (14) (5 versus 1). Thus, we should support a significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of head and neck cancer.
Two papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of head and neck cancer (16,23). One of them demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of head and neck cancer (23). But another paper did not show any significant association between them (16). The meta-analysis by Zhou Y (23) had a larger number of included studies than that by Liu P (16) (6 versus 1). Thus, we should support a significant association between HIF-1α rs11549467 (1790 G/A) polymorphism and the risk of head and neck cancer.
Glioma
A total of two meta-analysis papers explored the role of HIF in glioma (14,24) (Table S3). Among them, one paper explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (14), and another paper explored HIF-1α expression alone (24).
Risk
One paper explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of glioma (14). It demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of glioma (14).
Clinicopathological features
One paper explored the association of HIF-1α expression with the clinicopathological features of glioma (24). It demonstrated that HIF-1α expression was significantly associated with the tumor stage of glioma (24).
Oral cancer
A total of ten meta-analysis papers explored the role of HIF in oral cancer (14,16,18-20,25-29) (Table S4). Among them, four papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (18,27-29), three papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (14,19,25), two papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20), and one paper explored both HIF-1α and HIF-2α protein expressions (26).
Risk
Seven papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of oral cancer (14,18,19,25,27-29). All of them did not show any significant association between them (14,18,19,25,27-29).
Six papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of oral cancer (16,18,20,27-29). Four of them demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of oral cancer (16,20,27,28). But another two papers did not show any significant association between them (18,29). The meta-analyses by Sun X (27) and Yan Q (28) had a larger number of included studies than those by Liu P (16), Yang X (Plos One, 2013) (18), Yang X (Tumour Biol, 2014) (29), and Ye Y (20). Thus, we should support a significant association between HIF-1α rs11549467 (1790 G/A) polymorphism and the risk of oral cancer.
Prognosis
One paper explored the association of HIF-1α and HIF-2α protein expression with the prognosis of oral cancer (26). It demonstrated that neither HIF-1α nor HIF-2α protein expression was significantly associated with the survival of oral cancer (26).
Oropharyngeal cancer
Only one paper explored the role of HIF in oropharyngeal cancer (30) (Table S5). It explored the association of HIF-1α expression with the prognosis of oropharyngeal cancer (30). It demonstrated that HIF-1α expression was significantly associated with the survival of oropharyngeal cancer (30).
Nasopharyngeal cancer
Only one paper explored the role of HIF in nasopharyngeal cancer (31) (Table S6). It explored the association of HIF-1α expression with the risk and clinicopathological features of nasopharyngeal cancer (31). It demonstrated that HIF-1α expression was significantly associated with the risk, lymph node metastasis, and clinical stage of nasopharyngeal cancer (31).
Lung cancer
A total of 12 meta-analysis papers explored the role of HIF in lung cancer (11,12,14,16,18,23,25,28,32-35) (Table S7). Among them, 4 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (11,18,28,33), 3 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (12,14,25), 2 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,23), 1 paper explored both HIF-1α and HIF-2α protein expressions (32), and 2 papers explored HIF-1α protein expression alone (34,35).
Risk
Seven papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of lung cancer (11,12,14,18,25,28,33). Four of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of lung cancer (11,18,28,33). But another 3 papers did not show any significant association between them (12,14,25). The meta-analyses by He P (12), Hu X (25), Li Y (14), Yan Q (28), and Yang X (18) had a larger number of included studies than those by Anam MT (11) and Liao S (33) (3, 3, 3, 3, and 3 versus 2 and 2). Among the meta-analyses by He P (12), Hu X (25), Li Y (14), Yan Q (28), and Yang X (18), the included studies were completely identical (Table S8). Only the meta-analysis by Yang X employed a random-effect model (18). Thus, we should support a significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of lung cancer.
Six papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of lung cancer (11,16,18,23,28,33). All of them demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of lung cancer (11,16,18,23,28,33).
Clinicopathological features
One paper explored the association of HIF-1α protein expression with the clinicopathological features of lung cancer (34). It demonstrated that HIF-1α protein expression was significantly associated with the stage, pathological type, diameter, lymph node metastasis, and differentiation of lung cancer (34).
Prognosis
Three papers explored the association of HIF-1α protein expression with the prognosis of lung cancer (32,34,35). All of them demonstrated that HIF-1α protein expression was significantly associated with the survival of lung cancer (32,34,35).
One paper explored the association of HIF-2α protein expression with the prognosis of lung cancer (32). It demonstrated that HIF-2α protein expression was significantly associated with the survival of lung cancer (32).
Breast cancer
A total of 17 meta-analysis papers explored the role of HIF in breast cancer (11-14,16-20,22,23,25,28,36-39) (Table S9). Among them, 5 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (11,18,22,28,39), 7 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (12-14,17,19,25,36), 3 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20,23), and 2 papers explored HIF-1α protein expression alone (37,38).
Risk
Eleven papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of breast cancer (11,12,14,17-19,22,25,28,36,39). Three of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of breast cancer (14,18,39). But another 8 papers did not show any significant association between them (11,12,17,19,22,25,28,36). The meta-analyses by He P (12), Ren HT (36), Wu G (17), and Yan Q (28) had a larger number of included studies than those by Hu X (Tumour Biol, 2014) (25), Li Y (14), Yang X (18), Ye Y (19), Zhao T (22), and Anam MT (11) (6, 6, 6, and 6 versus 5, 5, 5, 3, 3, and 2). An abstract paper by Yin W did not report the number of included studies (39). Thus, we should not support any significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of breast cancer.
Eight papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of breast cancer (11,16,18,20,22,23,28,39). Two of them demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of breast cancer (11,22). But another 6 papers did not show any significant association between them (16,18,20,23,28,39). The meta-analysis by Yan Q (28) had a larger number of included studies than those by Liu P (16), Yang X (18), Zhou Y (23), Anam MT (11), Ye Y (20), and Zhao T (22) (4 versus 3, 3, 3, 2, 2, and 2). An abstract paper by Yin W did not report the number of included studies (39). Thus, we should not support any significant association between HIF-1α rs11549467 (1790 G/A) polymorphism and the risk of breast cancer.
Clinicopathological features
One paper explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the clinicopathological features of breast cancer (13). It did not show any significant association of HIF-1α rs11549465 (1772 C/T) polymorphism with the lymph node metastasis or histological grade of breast cancer (13).
One paper explored the association of HIF-1α protein expression with the clinicopathological features of breast cancer (37). It demonstrated that HIF-1α protein expression was significantly associated with the pathological differentiation, regional invasive extension, axillary lymph node status, and clinical stage of breast cancer (37).
Prognosis
Two papers explored the association of HIF-1α protein expression with the prognosis of breast cancer (37,38). Both of them demonstrated that HIF-1α protein expression was significantly associated with the survival of breast cancer (37,38).
Digestive cancer
A total of 33 meta-analysis papers explored the role of HIF in digestive cancer (Table S10). Among them, 6 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (15,27-29,40,41), 11 papers explored HIF-1α rs11549465 (1772 C/T)polymorphism alone (11-14,17-19,22,25,42,43), 3 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20,23), 1 paper explored both HIF-1α and HIF-2α protein expressions (44), 10 papers explored HIF-1α protein expression alone (45-54), and 2 papers explored HIF-2α protein expression alone (55,56).
Overall digestive cancer
A total of 8 meta-analysis papers explored the role of HIF in overall digestive cancer regardless of location of digestive cancer (17,20,27,29,40-43) (Table S10). Among them, 4 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (27,29,40,41), 3 papers explored HIF-1α rs11549465 (1772 C/T)polymorphism alone (17,42,43), and 1 paper explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (20).
Risk
Seven papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of overall digestive cancer (17,27,29,40-43). Four of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of overall digestive cancer (29,40,42,43). But another 3 papers did not show any significant association between them (17,27,41). The meta-analysis by Sun X (27) had a larger number of included studies than those by Yang X (29), Ni Z (40), Wu G (17), Xu JJ (Genet Mol Res, 2014) (41), Xu J (Genet Mol Res, 2014) (43), and Xu J (Genet Test Mol Biomarkers, 2013) (42) (13 versus 12, 10, 9, 8, 6, and 6). Thus, we should not support any significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of overall digestive cancer.
Five papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of overall digestive cancer (20,27,29,40,41). All of them demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of overall digestive cancer (20,27,29,40,41).
Esophageal cancer
A total of 5 meta-analysis papers explored the role of HIF in esophageal cancer (14,42,47,49,50) (Table S10). Among them, 2 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (14,42) and 3 papers explored HIF-1α protein expression alone (47,49,50).
Risk
Two papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of esophageal cancer (14,42). Both of them did not show any significant association between them (14,42).
Two papers explored the association of HIF-1α protein expression with the risk of esophageal cancer (47,50). Both of them demonstrated that HIF-1α protein expression was significantly associated with the risk of esophageal cancer (47,50).
Clinicopathological features
Three papers explored the association of HIF-1α protein expression with the clinicopathological features of esophageal cancer (47,49,50). All of them demonstrated that HIF-1α protein expression was significantly associated with the lymphoma node metastasis of esophageal cancer (47,49,50).
Prognosis
Two papers explored the association of HIF-1α protein expression with the prognosis of esophageal cancer (47,49). Both of them demonstrated that HIF-1α protein expression was significantly associated with the survival of esophageal cancer (47,49).
Gastric cancer
A total of 8 meta-analysis papers explored the role of HIF in gastric cancer (14,16,42,46,48,52,54) (Table S10). Among them, 2 papers explored HIF-1α rs11549465 (1772 C/T)polymorphism alone (14,42), 1 paper explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16), 4 papers explored HIF-1α protein expression alone (46,48,52,54), and 1 paper explored HIF-2α expression alone (56).
Risk
Two papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of gastric cancer (14,42). One of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of gastric cancer (42). But another paper did not show any significant association between them (14). The number of included studies was similar between the two meta-analysis papers by Li Y (14) and Xu J (42) (1 versus 1). The included study was also identical between the two meta-analysis papers (Table S11). After learning the results from the original study (Li K, et al. Biochem Genet, 2009) (57), we should not support any significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of gastric cancer.
One paper explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of gastric cancer (16). It demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of gastric cancer (16).
Clinicopathological features
Three papers explored the association of HIF-1α protein expression with the clinicopathological features of gastric cancer (46,48,54). All of them demonstrated that HIF-1α protein expression was significantly associated with the depth of invasion, lymphatic invasion, vascular invasion, and TNM stage of gastric cancer (46,48,54).
One paper explored the association of HIF-2α protein expression with the clinicopathological features of gastric cancer (56). It demonstrated that HIF-2α protein expression was significantly associated with the tumor infiltration, lymphatic metastasis, and TNM stage of gastric cancer (56).
Prognosis
Four papers explored the association of HIF-1α protein expression with the prognosis of gastric cancer (46,48,52,54). All of them demonstrated that HIF-1α protein expression was significantly associated with the survival of gastric cancer (46,48,52,54).
One paper explored the association of HIF-2α protein expression with the prognosis of gastric cancer (56). It demonstrated that HIF-2α protein expression was significantly associated with the survival of gastric cancer (56).
Colorectal cancer
A total of 15 meta-analysis papers explored the role of HIF in colorectal cancer (11-16,18,19,22,25,27-29,42,44) (Table S10). Among them, 4 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (15,27-29), 9 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (11-14,18,19,22,25,42), 1 paper explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16), and 1 paper explored both HIF-1α and HIF-2α protein expressions (44).
Risk
Twelve papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of colorectal cancer (11,12,14,15,18,19,22,25,27-29,42,55). All of them did not show any significant association between them (11,12,14,15,18,19,22,25,27-29,42).
Four papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of colorectal cancer (16,27-29). All of them did not show any significant association between them (16,27-29).
Clinicopathological features
One paper explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the clinicopathological features of colorectal cancer (13). It did not show any significant association of HIF-1α rs11549465 (1772 C/T) polymorphism with the lymph node metastasis and histological grade of colorectal cancer (13).
One paper explored the association of HIF-1α protein expression with the clinicopathological features of colorectal cancer (44). It demonstrated that HIF-1α protein expression was significantly associated with the Dukes’ stages, lymph node status, depth of invasion, metastasis, and UICC stage of colorectal cancer, but not the differentiation grade (44).
One paper explored the association of HIF-2α protein expression with the clinicopathological features of colorectal cancer (44). It demonstrated that HIF-2α protein expression was significantly associated with the differentiation grade of colorectal cancer, but not the Dukes’ stages, lymph node status, or depth of invasion (44).
Prognosis
One paper explored the association of HIF-1α protein expression with the prognosis of colorectal cancer (44). It demonstrated that HIF-1α protein expression was significantly associated with the survival of colorectal cancer (44).
One paper explored the association of HIF-2α protein expression with the prognosis of colorectal cancer (44). It demonstrated that HIF-2α protein expression was significantly associated with the survival of colorectal cancer (44).
Pancreatic cancer
A total of 8 meta-analysis papers explored the role of HIF in pancreatic cancer (12,14,16,23,27-29,51) (Table S10). Among them, 2 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (27,29), 2 papers explored HIF-1α rs11549465 (1772 C/T)polymorphism alone (12,14), 3 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,23,28), and 1 paper explored HIF-1α protein expression alone (51).
Risk
Four papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of pancreatic cancer (12,14,27,29). All of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of pancreatic cancer (12,14,27,29).
Five papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of pancreatic cancer (16,23,27-29). All of them demonstrated that HIF-1α rs11549467 (1790 G/A)polymorphism was significantly associated with the risk of pancreatic cancer (16,23,27-29).
Clinicopathological features
One paper explored the association of HIF-1α protein expression with the clinicopathological features of pancreatic cancer (51). It demonstrated that HIF-1α protein expression was significantly associated with the lymph node metastasis and tumor stage of pancreatic cancer, but not the tumor size (51).
Prognosis
One paper explored the association of HIF-1α protein expression with the prognosis of pancreatic cancer (51). It demonstrated that HIF-1α protein expression was significantly associated with the survival of pancreatic cancer (51).
Hepatocellular carcinoma
A total of 5 meta-analysis papers explored the role of HIF in hepatocellular carcinoma (14,16,45,53,55) (Table S10). Among them, 1 paper explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (14), 1 paper explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16), 2 papers explored HIF-1α protein expression alone (45,53), and 1 paper explored HIF-2α protein expression alone (55).
Risk
One paper explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of hepatocellular carcinoma (14). It did not show any significant association between them (14).
One paper explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of hepatocellular carcinoma (16). It demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of hepatocellular carcinoma (16).
Clinicopathological features
One paper explored the association of HIF-1α protein expression with the clinicopathological features of hepatocellular carcinoma (46). It demonstrated that HIF-1α protein expression was significantly associated with the vascular invasion of hepatocellular carcinoma, but not the tumor size or differentiation, liver cirrhosis, or capsule formation (46).
One paper explored the association of HIF-2α protein expression with the clinicopathological features of hepatocellular carcinoma (55). It demonstrated that HIF-2α protein expression was significantly associated with the vein invasion, histological grade, and capsule infiltration of hepatocellular carcinoma, but not the tumor size or liver cirrhosis (55).
Prognosis
Two papers explored the association of HIF-1α protein expression with the prognosis of hepatocellular carcinoma (46,53). Both of them demonstrated that HIF-1α protein expression was significantly associated with the survival of hepatocellular carcinoma (46,53).
One paper explored the association of HIF-2α protein expression with the prognosis of hepatocellular carcinoma (55). It did not show any significant association between HIF-2α protein expression and the survival of hepatocellular carcinoma (55).
Urinary cancer
A total of 15 meta-analysis papers explored the role of HIF in urinary cancer (11,12,14,16-20,22,23,25,28,58-60) (Table S12). Among them, 5 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (11,18,22,28,59), 5 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (12,14,17,19,25), 3 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20,23), 1 paper explored both HIF-1α and HIF-2α protein expressions (58), and 1 paper explored HIF-1α protein expression alone (60).
Overall urinary cancer
One meta-analysis paper explored the association of HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms with the risk of overall urinary cancer (59) (Table S12). It demonstrated that neither of them was significantly associated with the risk of overall urinary cancer (59).
Prostate cancer
A total of 13 meta-analysis papers explored the role of HIF in prostate cancer (11,12,14,16-20,22,23,25,28,59) (Table S12). Among them, 5 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (11,18,22,28,59), 5 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (11,18,22,28,59), and 3 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20,23).
Risk
Ten papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of prostate cancer (11,12,14,17-19,22,25,59). Five of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of prostate cancer (14,18,19,22,25). Another 5 papers did not show any significant association between them (11,12,17,28,59). The meta-analyses by Anam MT (11), He P (12), Li D (59), Wu G (17), and Yan Q (28) had a larger number of included studies than those by Hu X (25), Yang X (18), Ye Y (19), Li Y (14), and Zhao T (22) (6, 6, 6, 6, and 6 versus 5, 5, 5, 4, and 4). Thus, we should not support any significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of prostate cancer.
Eight papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of prostate cancer (11,16,18,20,22,23,28,59). One of them demonstrated that HIF-1α rs11549467 (1790 G/A)polymorphism was significantly associated with the risk of prostate cancer (59). Another 7 papers did not show any significant association between them (11,16,18,20,22,23,28). The meta-analysis by Li D (59) had a larger number of included studies than those by Anam MT (11), Liu P (16), Yan Q (28), Ye Y (20), Yang X (18), Zhou Y (23), and Zhao T (22) (4 versus 3, 3, 3, 3, 3, 3, and 2). Thus, we should support a significant association between HIF-1α rs11549467 (1790 G/A) polymorphism and the risk of prostate cancer.
Renal cancer
A total of 13 meta-analysis papers explored the role of HIF in renal cancer (11,12,14,16,17,19,20,23,25,28,58-60) (Table S12). Among them, 3 papers explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (11,28,59), 5 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (12,14,17,19,25), 3 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20,23), 1 paper explored both HIF-1α and HIF-2α nuclear and cytoplasmic expressions (58), and 1 paper explored HIF-1α protein expression alone (60).
Risk
Eight papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of renal cancer (11,12,14,17,19,25,28,59). Two of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of renal cancer (11,12). Another 6 papers did not show any significant association between them (14,17,19,25,28,59). The meta-analyses by Hu X (25), Li D (59), Wu G (17), and Yan Q (28) had a larger number of included studies than those by Anam MT (11), He P (12), Ye Y (19), and Li Y (14) (4, 4, 4, and 4 versus 3, 3, 3, and 2). Thus, we should not support any significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of renal cancer.
Six papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of renal cancer (11,16,20,23,28,59). Three of them demonstrated that HIF-1α rs11549467 (1790 G/A) polymorphism was significantly associated with the risk of renal cancer (11,23,28). Another 3 papers did not show any significant association between them (16,19,59). The meta-analyses by Anam MT (11), Li D (59), and Yan Q (28) had a larger number of included studies than those by Liu P (16), Zhou Y (23), and Ye Y (20) (4, 4, and 4 versus 3, 3, and 2). The included studies were completely identical among the 3 meta-analyses by Anam MT (11), Li D (59), and Yan Q (28) (Table S13). Notably, some statistical results (AA + AG vs. GG and A allele vs. G allele) were completely identical among them (11,28,59). However, the meta-analyses by Anam MT (11) and Yan Q (28) had more statistical results (AA vs. GG, GA vs. GG, and AA vs. GA + GG) than that by Li D (59). Thus, we should support a significant association between HIF-1α rs11549467 (1790 G/A) polymorphism and the risk of renal cancer.
Clinicopathological features
One paper explored the association of HIF-1α protein expression with the clinicopathological features of renal cancer (60). It demonstrated that HIF-1α protein expression was significantly associated with the lymph node metastasis and clinical and pathological stage of renal cancer (60).
Prognosis
One paper explored the association of HIF-1α and HIF-2α nuclear and cytoplasmic expressions with the prognosis of renal cancer (58). It demonstrated that neither HIF-1α nor HIF-2α nuclear and cytoplasmic expression was significantly associated with the survival of renal cancer (58).
Bladder cancer
A total of 3 meta-analysis papers explored the role of HIF in bladder cancer (14,16,25) (Table S12). Among them, 2 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (14,25), and 1 paper explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16).
Risk
Two papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of bladder cancer (14,25). Neither of them demonstrated any significant association between HIF-1α rs11549465 (1772 C/T)polymorphism and the risk of bladder cancer (14,25).
One paper explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of bladder cancer (16). It did not show any significant association between them (16).
Gynecological cancer
A total of 12 meta-analysis papers explored the role of HIF in gynecological cancer (12-14,16,19,20,25,28,61-64) (Table S14). Among them, 1 paper explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (28), 8 papers explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (12-14,19,25,64), 2 papers explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16,20), and 1 paper explored HIF-1α protein expression alone (61-63,65).
Overall gynecological cancer
A total of 3 meta-analysis papers explored the role of HIF in overall gynecological cancer (14,16,65) (Table S14). Among them, 1 paper explored HIF-1α rs11549465 (1772 C/T) polymorphism alone (14), 1 paper explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (16), and 1 paper explored HIF-1α protein expression alone (65).
Risk
One paper explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of overall gynecological cancer (14). It demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of overall gynecological cancer (14).
One paper explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of overall gynecological cancer (16). It did not show any significant association between them (16).
Clinicopathological features
One paper explored the association of HIF-1α protein expression with the clinicopathological features of overall gynecological cancer (65). It demonstrated that HIF-1α protein expression was significantly associated with the pathological and histological type, FIGO stage, and lymph node metastasis of overall gynecological cancer (65).
Prognosis
One paper explored the association of HIF-1α protein expression with the prognosis of overall gynecological cancer (65). It demonstrated that HIF-1α protein expression was significantly associated with the survival of overall gynecological cancer (65).
Ovarian cancer
A total of 3 meta-analysis papers explored the role of HIF in ovarian cancer (62,63,65). All of them explored HIF-1α protein expression alone (62,63,65) (Table S14).
Risk
One paper explored the association of HIF-1α protein expression with the risk of ovarian cancer (63). It demonstrated that HIF-1α protein expression was significantly associated with the risk of ovarian cancer (63).
Clinicopathological features
Three papers explored the association of HIF-1α protein expression with the lymph node metastasis of ovarian cancer (62,63,65). All of them demonstrated that HIF-1α protein expression was significantly associated with the lymph node metastasis of ovarian cancer (62,63,65).
Three papers explored the association of HIF-1α protein expression with the pathological type of ovarian cancer (62,63,65). Two of them demonstrated HIF-1α protein expression was significantly associated with the pathological type of ovarian cancer (62,65). But another paper did not show any significant association between them (63). The meta-analyses by Jin Y (Tumour Biol, 2014) (62) and Jin Y (PLoS One, 2015) (65) had a larger number of included studies than that by Sun C (63) (13 and 13 versus 4). Thus, we should support a significant association between HIF-1α protein expression and the pathological type of ovarian cancer.
Two papers explored the association of HIF-1α protein expression with the FIGO stage of ovarian cancer (62,65). Both of them demonstrated that HIF-1α protein expression was significantly associated with the FIGO stage of ovarian cancer (62,65).
Prognosis
Two papers explored the association of HIF-1α protein expression with the prognosis of ovarian cancer (62,65). Both of them demonstrated that HIF-1α protein expression was significantly associated with the survival of ovarian cancer (62,65).
Cervical cancer
A total of 10 meta-analysis papers explored the role of HIF in cervical cancer (12,13,18-20,25,28,61,64,65) (Table S14). Among them, 1 paper explored both HIF-1α rs11549465 (1772 C/T) and rs11549467 (1790 G/A) polymorphisms (28), 6 papers explored HIF-1α rs11549465 (1772 C/T)polymorphism alone (12,13,18,19,25,64), 1 paper explored HIF-1α rs11549467 (1790 G/A) polymorphism alone (20), and 2 papers explored HIF-1α protein expression alone (61,65).
Risk
Six papers explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the risk of cervical cancer (12,18,19,25,28,64). Five of them demonstrated that HIF-1α rs11549465 (1772 C/T) polymorphism was significantly associated with the risk of cervical cancer (12,18,25,28,64). But another paper did not show any significant association between them (19). The meta-analysis by Zhu J (64) had a larger number of included studies than those by He P (12), Hu X (25), Yan Q (28), Yang X (18), and Ye Y (19) (4 versus 3, 3, 3, 3, and 3). Thus, we should support a significant association between HIF-1α rs11549465 (1772 C/T) polymorphism and the risk of cervical cancer.
Two papers explored the association of HIF-1α rs11549467 (1790 G/A) polymorphism with the risk of cervical cancer (20,28). Neither of them showed any significant association between HIF-1α rs11549467 (1790 G/A) polymorphism and the risk of cervical cancer (20,28).
Clinicopathological features
One paper explored the association of HIF-1α rs11549465 (1772 C/T) polymorphism with the lymph node metastasis of cervical cancer (13). It did not show any significant association between them (13).
Two papers explored the association of HIF-1α protein expression with the FIGO stage of cervical cancer (61,65). Both of them demonstrated that HIF-1α protein expression was significantly associated with the FIGO stage of cervical cancer (61,65).
Two papers explored the association of HIF-1α protein expression with the histological type and lymph node metastasis of cervical cancer (61,65). One of them demonstrated that HIF-1α protein expression was significantly associated with the histological type and lymph node metastasis of cervical cancer (65). But another paper did not show any significant association between them (61). As for the histological type, the meta-analysis by Jin Y (65) had a larger number of included studies than that by Huang M (61) (6 versus 4). As for the lymph node metastasis, the meta-analysis by Jin Y (65) had a larger number of included studies than that by Huang M (61) (8 versus 5). Thus, we should support a significant association between HIF-1α protein expression and the histological type and lymph node metastasis of cervical cancer.
Prognosis
Two papers explored the association of HIF-1α protein expression with the prognosis of cervical cancer (61,65). Both of them demonstrated that HIF-1α protein expression was significantly associated with the survival of cervical cancer (61,65).
Endometrial cancer
Only one paper explored the association of HIF-1α protein expression with the clinicopathological features and prognosis of endometrial cancer (65) (Table S14). It demonstrated that HIF-1α protein expression was significantly associated with the pathological and histological type, FIGO stage, and lymph node metastasis of endometrial cancer, but not the survival (65).
Osteosarcoma
Only one paper explored the association of HIF-1α protein expression with the clinicopathological features and prognosis of osteosarcoma (66) (Table S15). It demonstrated that HIF-1α protein expression was significantly associated with the metastasis, pathologic and tumor grade, and survival of osteosarcoma, but not the histopathology, tumor size, or tumor site (66).
Conclusions
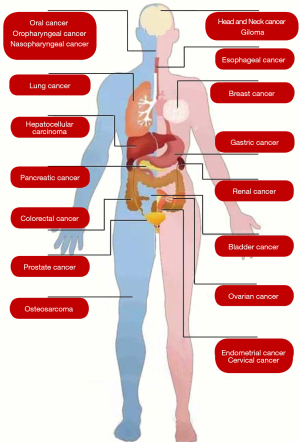
Based on our systematic search strategy, numerous meta-analyses have explored the role of HIF gene polymorphism and protein expression in various human cancers, including head and neck cancer, glioma, oral cancer, oropharyngeal cancer, nasopharyngeal cancer, lung cancer, breast cancer, esophageal cancer, gastric cancer, colorectal cancer, pancreatic cancer, hepatocellular carcinoma, prostate cancer, renal cancer, bladder cancer, ovarian cancer, cervical cancer, endometrial cancer, and osteosarcoma (Figure 2).
Based on the current evidence, major findings were summarized in Table 1.
Table 1
| Cancer | Risk | Lymph node metastasis/tumor stage | Survival | |||||
|---|---|---|---|---|---|---|---|---|
| HIF-1α rs11549465 (1772 C/T) polymorphism | HIF-1α rs11549467 (1790 G/A) polymorphism | HIF-1α expression | HIF-2α expression | HIF-1α expression | HIF-2α expression | |||
| Head and neck cancer | Y | Y | ||||||
| Glioma | Y | Y | ||||||
| Oral cancer | N | Y | N | N | ||||
| Oropharyngeal cancer | Y | |||||||
| Nasopharyngeal cancer | Y | |||||||
| Lung cancer | Y | Y | Y/Y | Y | Y | |||
| Breast cancer | N | N | Y | Y | ||||
| Esophageal cancer | N | Y | Y | |||||
| Gastric cancer | N | Y | Y/Y | Y/Y | Y | Y | ||
| Colorectal cancer | N | N | Y/Y | N/N | Y | Y | ||
| Pancreatic cancer | Y | Y | Y/Y | Y | ||||
| Hepatocellular carcinoma | N | Y | Y | N | ||||
| Prostate cancer | N | Y | ||||||
| Renal cancer | N | Y | Y/Y | N | N | |||
| Bladder cancer | N | N | ||||||
| Ovarian cancer | Y/Y | Y | ||||||
| Cervical cancer | Y | N | Y | |||||
| Endometrial cancer | N | |||||||
| Osteosarcoma | Y | Y | ||||||
Y, There is a significant correlation; N, There is no significant correlation.
First, the evidence regarding the association of HIF-1α gene polymorphism with risk of cancer suggested the following: (I) both HIF-1α rs11549465 (1772 C/T) and HIF-1α rs11549467 (1790 G/A) polymorphisms should be associated with the risk of head and neck cancer and lung cancer; (II) HIF-1α rs11549465 (1772 C/T) polymorphism, rather than HIF-1α rs11549467 (1790 G/A) polymorphism, should be associated with the risk of cervical cancer; (III) HIF-1α rs11549467 (1790 G/A) polymorphism, rather than HIF-1α rs11549465 (1772 C/T) polymorphism, should be associated with the risk of oral cancer, gastric cancer, hepatocellular carcinoma, prostate cancer, and renal cancer; and (IV) neither HIF-1α rs11549465 (1772 C/T) nor HIF-1α rs11549467 (1790 G/A) polymorphism should be associated with the risk of breast cancer, colorectal cancer, and bladder cancer.
Second, the evidence regarding the association of HIF-1α protein expression with the lymph node metastasis of cancer suggested the following: (I) both HIF-1α and HIF-2α expression were associated with the lymph node metastasis of gastric cancer; and (II) HIF-1α expression, rather than HIF-2α expression, was associated with the lymph node metastasis of colorectal cancer.
Third, the evidence regarding the association of HIF-1α protein expression alone with the lymph node metastasis of cancer suggested that HIF-1α expression was associated with the lymph node metastasis of glioma, nasopharyngeal cancer, lung cancer, breast cancer, esophageal cancer, gastric cancer, pancreatic cancer, renal cancer, ovarian cancer, and osteosarcoma.
Fourth, the evidence regarding the association of HIF-1α protein expression with the survival of cancer suggested the following: (I) both HIF-1α and HIF-2α expressions were associated with the survival of lung cancer, gastric cancer, and colorectal cancer; (II) HIF-1α expression, rather than HIF-2α expression, was associated with the survival of hepatocellular carcinoma; and (III) neither HIF-1α nor HIF-2α expression was associated with the survival of renal cancer.
Fifth, the evidence regarding the association of HIF-1α protein expression alone with the survival of cancer suggested that HIF-1α expression was associated with the survival of oropharyngeal cancer, breast cancer, esophageal cancer, pancreatic cancer, ovarian cancer, cervical cancer, and osteosarcoma, but not that of endometrial cancer.
Collectively, the impact of HIFs on the risk, clinicopathological features, and survival of various human cancers should be heterogeneous. The potential explanation might be attributed to the heterogeneity in the cancer biological behavior and effect of hypoxia across the different types of human cancers. Further studies should uncover the potential mechanisms.
Table S1
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Anam MT | Biomark Res [2015] | Bangladesh | PubMed, PubMed Central, Google Scholar | 2014.12 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 22 | Risk: |
| TT vs. CC: OR =1.52, 95% CI: 0.73–3.18, P=0.2648 | ||||||||
| CT vs. CC: OR =1.23, 95% CI: 1.00–1.53, P=0.0536 | ||||||||
| TT + CT vs. CC: OR =1.30, 95% CI: 1.06–1.59, P=0.0115 | ||||||||
| TT vs. CT + CC: OR =1.64, 95% CI: 0.94–2.85, P=0.0832 | ||||||||
| T allele vs. C allele: OR =1.32, 95% CI: 1.07–1.63, P=0.0098 | ||||||||
| Overall cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 19 | Risk: | |||||
| AA vs. GG: OR =5.10, 95% CI: 3.12–8.33, P<0.0001 | ||||||||
| GA vs. GG: OR =1.74, 95% CI: 1.20–2.52, P=0.0033 | ||||||||
| AA vs. GA + GG: OR =3.79, 95% CI: 2.34–6.15, P<0.0001 | ||||||||
| AA + GA vs. GG: OR =1.82, 95% CI: 1.26–2.62, P=0.0014 | ||||||||
| A allele vs. G allele: OR =1.82, 95% CI: 1.31–2.52, P=0.0003 | ||||||||
| He P | PLoS One [2013] | China | PubMed, Embase, CNKI | 2013.8.23 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 36 | Dominant model (TT + CT vs. CC): OR =1.23, 95% CI: 1.03–1.47 | |||||||
| 26 | Recessive model (TT vs. CT + CC): OR =2.51, 95% CI: 1.54–4.09 | |||||||
| 25 | Homozygote comparison (TT vs. CC): OR =2.02, 95% CI: 1.21–3.39 | |||||||
| 36 | Heterozygote comparison (CT vs. CC): OR =1.16, 95% CI: 0.97–1.38 | |||||||
| Hu X | Tumour Biol [2013] | China | PubMed, Embase, CNKI | 2013.2 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 15 | Lymph node metastasis: |
| OR =1.38, 95% CI: 1.13–1.68, P=0.002 | ||||||||
| 7 | Distant metastasis: | |||||||
| OR =1.39, 95% CI: 0.96–2.02, P=0.082 | ||||||||
| 9 | Tumor size: | |||||||
| T2–4 vs. T1: OR =1.09, 95% CI: 0.83–1.45, P=0.530 | ||||||||
| T3–4 vs. T1–2: OR =1.29, 95% CI: 0.93–1.80, P=0.128 | ||||||||
| 5 | Stage: | |||||||
| OR =0.93, 95% CI: 0.66–1.31, P=0.43 | ||||||||
| 9 | Histological grade: | |||||||
| Grades G3 vs. G1: OR =1.07, 95% CI: 0.71–1.60, P=0.759 | ||||||||
| Grades G3 vs. G2: OR =1.51, 95% CI: 1.08–2.13, P=0.017 | ||||||||
| Grades G2 vs. G1: OR =0.67, 95% CI: 0.46–0.97, P=0.035 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 8 | Lymph node metastasis: | ||||||
| OR =1.33, 95% CI: 1.00–1.78, P=0.050 | ||||||||
| 4 | Distant metastasis: | |||||||
| OR =0.97, 95% CI: 0.58–1.62, P=0.893 | ||||||||
| 5 | Tumor size: | |||||||
| T2–4 vs. T1: OR =1.04, 95% CI: 0.65–1.65, P=0.871 | ||||||||
| T3–4 vs. T1–2: OR =1.64, 95% CI: 1.04–2.58, P=0.033 | ||||||||
| 4 | Stage: | |||||||
| OR =1.00, 95% CI: 0.65–1.52, P=0.987 | ||||||||
| 5 | Histological grade: | |||||||
| Grades G3 vs. G1: OR =0.93, 95% CI: 0.56–1.55, P=0.789 | ||||||||
| Grades G3 vs. G2: OR =1.12, 95% CI: 0.73–1.70, P=0.609 | ||||||||
| Grades G2 vs. G1: OR =0.88, 95% CI: 0.57–1.36, P=0.556 | ||||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 28 | Risk: |
| TT vs. CC: OR =2.15, 95% CI: 1.19–3.88, P=0.011 | ||||||||
| CT vs. CC: OR =1.15, 95% CI: 0.96–1.36, P=0.127 | ||||||||
| TT/CT vs. CC: OR =1.19, 95% CI: 0.99–1.42, P=0.071 | ||||||||
| TT vs. CT/CC: OR =2.21, 95% CI: 1.60–3.05, P=0.010 | ||||||||
| T allele vs. C allele: OR =1.20, 95% CI: 1.01–1.44, P=0.043 | ||||||||
| Liu J | Gene [2013] | China | PubMed, Embase | 2012.3 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 8 | Allele: OR =1.177, 95% CI=1.011–1.369, P=0.035 | |||||||
| 7 | Genotype: OR =0.975, 95% CI=0.868–1.055, P=0.373 | |||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |||||||
| 6 | Allele: OR =1.254, 95% CI=0.77–2.043, P=0.362 | |||||||
| 5 | Genotype: OR =0.736, 95% CI=0.595–0.910, P=0.005 | |||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Overall cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 26 | Risk: |
| AA vs. GG: OR =4.37, 95% CI: 2.61–7.33, p<0.001 | ||||||||
| GA vs. GG: OR =1.39, 95% CI: 1.06–1.82, P=0.017 | ||||||||
| AA + GA vs. GG: OR =1.46, 95% CI: 1.11–1.92, P=0.007 | ||||||||
| AA vs. GA + GG: OR =3.87, 95% CI: 2.32–6.46, P<0.001 | ||||||||
| A allele vs. G allele: OR =1.49, 95% CI: 1.15–1.95, P=0.003 | ||||||||
| Wu G | Tumour Biol [2014] | China | PubMed, Embase, Google Scholar, Wanfang | 2013.6.10 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 38 | Risk: |
| TT + CT vs. CC: OR =1.18, 95% CI: 1.00–1.38, P=0.048 | ||||||||
| 35 | TT vs. CT + CC: OR =1.22, 95% CI: 1.05–1.41, P=0.01 | |||||||
| Yang X | PLoS One [2013] | China | PubMed, Embase | 2013.6.26 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 34 | Risk: |
| TT vs. CC: OR =2.45, 95% CI: 1.52–3.96 | ||||||||
| CT vs. CC: OR =1.15, 95% CI: 0.92–1.45 | ||||||||
| TT + CT vs. CC: OR =1.27, 95% CI: 1.05–1.55 | ||||||||
| TT vs. CT + CC: OR =3.18, 95% CI: 1.92–5.29 | ||||||||
| T allele vs. C allele: OR =1.42, 95% CI: 1.18–1.70 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 24 | Risk: | ||||||
| AA vs. GG: OR =4.74, 95% CI: 1.78–12.6 | ||||||||
| GA vs. GG: OR =1.35, 95% CI: 0.82–2.21 | ||||||||
| AA + GA vs. GG: OR =1.65, 95% CI: 1.05–2.60 | ||||||||
| AA vs. GA + GG: OR =4.39, 95% CI: 1.61–11.9 | ||||||||
| A allele vs. G allele: OR =1.83, 95% CI: 1.13–2.96 | ||||||||
| Ye Y | Cancer Invest [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 29 | Risk: |
| TT + CT vs. CC: OR =1.28, 95% CI: 1.06–1.54, P=0.009 | ||||||||
| Ye Y | Tumori [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Overall cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 21 | Risk: |
| TT + CT vs. CC: OR =1.79, 95% CI: 1.12–2.86, P=0.01 | ||||||||
| Zhang Q | PLoS One [2013] | China | PubMed, Embase | 2012.12.1 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 15 | TT+CT vs. CC: OR =1.39, 95% CI: 1.13–1.71, P=0.002 | |||||||
| 5 | TT vs. CT+CC: OR =1.93, 95% CI: 0.86–4.36, P=0.11 | |||||||
| 15 | T allele vs. C allele: OR =1.36, 95% CI: 1.12–1.64, P=0.002 | |||||||
| Zhao T | J Exp Clin Cancer Res [2009] | China | PubMed | 2009.6 | Overall cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 18 | Risk: |
| T allele vs. C allele: OR =1.29, 95% CI: 1.01–1.65, P=0.04 | ||||||||
| TT vs. CT + CC: OR =2.18, 95% CI: 1.32–3.62, P=0.003 | ||||||||
| TT + CT vs. CC: OR =1.19, 95% CI: 0.88–1.59, P=0.26 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 12 | Risk: | ||||||
| A allele vs. G allele: OR =1.61, 95% CI: 0.75–3.45, P=0.22 | ||||||||
| AA + GA vs. GG: OR =1.56, 95% CI: 0.66–3.65, P=0.31 | ||||||||
| Zhou Y | Cancer Cell Int [2014] | China | PubMed, Embase, CNKI | 2013.12.13 | Overall cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |
| 25 | AA + GA vs. GG: OR =1.85, 95% CI: 1.27–2.69 | |||||||
| 26 | AA vs. GA + GG: OR =5.69, 95% CI: 3.87–8.37 | |||||||
| 12 | AA vs. GG: OR =6.63, 95% CI: 4.49–9.79 | |||||||
| 11 | GA vs. GG: OR =2.39, 95% CI: 1.53–3.75 |
Table S2
| First author | Journal (year) | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| He P | PLoS One [2013] | China | PubMed, Embase, CNKI | 2013.8.23 | Head and neck cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 5 | Dominant model (TT + CT vs. CC): OR =1.20, 95% CI: 0.87–1.67 | |||||||
| 4 | Recessive model (TT vs. CT + CC): OR =11.29, 95% CI: 1.24–103.02 | |||||||
| 3 | Homozygote comparison (TT vs. CC): OR =2.24, 95% CI: 1.14–4.39 | |||||||
| 5 | Heterozygote comparison (CT vs. CC): OR =1.03, 95% CI: 0.69–1.62 | |||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Head and neck squamous cell carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: |
| CT vs. CC: OR =1.81, 95% CI: 0.73–4.51, P=0.199 | ||||||||
| TT /CT vs. CC: OR =1.81, 95% CI: 0.73–4.51, P=0.199 | ||||||||
| T allele vs. C allele: OR =1.73, 95% CI: 0.72–4.15, P=0.217 | ||||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Head and neck squamous cell carcinoma | HIF-1α rs11549467 (1790 G/A) polymorphism | 1 | Risk: |
| GA vs. GG: OR =0.88, 95% CI: 0.26–3.00, P=0.838 | ||||||||
| AA + GA vs. GG: OR =0.88, 95% CI: 0.26–3.00, P=0.838 | ||||||||
| A allele vs. G allele: OR =0.88, 95% CI: 0.27–2.94, P=0.841 | ||||||||
| Zhou Y | Cancer Cell Int [2014] | China | PubMed, Embase, CNKI | 2013.12.13 | Head and neck cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |
| 6 | AA + GA vs. GG: OR =3.57, 95% CI: 0.98–12.99 | |||||||
| 3 | AA vs. GA + GG: OR =58, 95% CI: 1.75–1,924.88 | |||||||
| 3 | AA vs. GG: OR =101.38, 95% CI: 22.09–65.29 | |||||||
| 3 | GA vs. GG: OR =12.53, 95% CI: 2.42–64.76 |
Table S3
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Glioma | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: |
| TT vs. CC: OR =2.23, 95% CI: 0.20–24.92, P=0.514 | ||||||||
| CT vs. CC: OR =2.15, 95% CI: 1.08–4.29, P=0.030 | ||||||||
| TT/CT vs. CC: OR =2.16, 95% CI: 1.10–4.21, P=0.025 | ||||||||
| TT vs. CT/CC: OR =2.01, 95% CI: 0.18–22.45, P=0.569 | ||||||||
| T allele vs. C allele: OR =2.05, 95% CI: 1.09–3.83, P=0.025 | ||||||||
| Liu Q | Int J Clin Exp Med [2015] | China | PubMed, Embase, Wanfang, CNKI | 2015 | Glioma | HIF-1α expression | 24 | IV + III vs. II+I: |
| OR =8.59, 95% CI: 6.56–11.24, P<0.00001 | ||||||||
| 14 | IV vs. III: | |||||||
| OR =2.51, 95% CI: 1.43–4.42, P=0.001 | ||||||||
| 11 | IV vs. II: | |||||||
| OR =9.18, 95% CI: 5.18–16.28, P<0.00001 | ||||||||
| 9 | IV vs. I: | |||||||
| OR = 24.23, 95% CI: 12.21–48.09, P<0.00001 | ||||||||
| 12 | III vs. II: | |||||||
| OR =4.59, 95% CI: 2.96–7.12, P<0.00001 | ||||||||
| 10 | III vs. I: | |||||||
| OR =13.34, 95% CI: 7.53–23.62, P<0.00001 | ||||||||
| 11 | II vs. I: | |||||||
| OR =4.19, 95% CI: 2.59–6.77, P<0.00001 |
Table S4
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Hu X | Tumour Biol [2014] | China | PubMed, Embase, CNKI | 2013.7 | Oral cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| T allele vs. C allele: OR =2.52, 95% CI: 0.71–8.98 | ||||||||
| TT vs. CC: OR =1.97, 95% CI: 0.72–5.39 | ||||||||
| CT vs. CC: OR =0.92, 95% CI: 0.44–1.89 | ||||||||
| TT + CT vs. CC: OR =1.06, 95% CI: 0.64–1.76 | ||||||||
| TT vs. CT + CC: OR =22.82, 95% CI: 0.28–1,887.72 | ||||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Oral squamous cell carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: |
| TT vs. CC: OR =6.14, 95% CI: 0.25–151.49, P=0.267 | ||||||||
| CT vs. CC: OR =1.28, 95% CI: 0.69–2.38, P=0.432 | ||||||||
| TT/CT vs. CC: OR =1.35, 95% CI: 0.73–2.49, P=0.334 | ||||||||
| TT vs. CT/CC: OR =6.01, 95% CI: 0.24–148.26, P=0.273 | ||||||||
| T allele vs. C allele: OR =1.41, 95% CI: 0.78–2.56, P=0.257 | ||||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Oral squamous cell carcinoma | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: |
| AA vs. GG: OR =13.32, 95% CI: 1.57–112.75, P=0.017 | ||||||||
| GA vs. GG: OR =2.96, 95% CI: 1.05–8.31, P=0.039 | ||||||||
| AA + GA vs. GG: OR =3.15, 95% CI: 1.05–9.47, P=0.041 | ||||||||
| AA vs. GA + GG: OR =10.70, 95% CI: 1.25–91.51, P=0.030 | ||||||||
| A allele vs. G allele: OR =3.09, 95% CI: 1.07–8.93, P=0.038 | ||||||||
| Qian J | Tumour Biol [2016] | China | PubMed, Web of Knowledge, Web of Science | 2016.1.12 | Oral squamous cell carcinoma | HIF-1α expression | 12 | OS: |
| RR =1.18, 95% CI: 0.66–2.11 | ||||||||
| HIF-2α expression | 2 | OS: | ||||||
| RR =1.40; 95% CI: 0.93–2.09 | ||||||||
| Sun X | World J Gastroenterol [2015] | China | PubMed, Embase, CNKI | 2013.7.15 | Oral cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| CT vs. CC: OR =0.917, 95% CI: 0.444–1.895 | ||||||||
| TT + CT vs. CC: OR =1.063, 95% CI: 0.643–1.757 | ||||||||
| T allele vs. C allele: OR =2.517, 95% CI: 0.705–8.980 | ||||||||
| Oral cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 4 | Risk: | |||||
| CT vs. CC: OR =3.165, 95% CI: 1.264–7.924 | ||||||||
| TT + CT vs. CC: OR =7.919, 95% CI: 1.582–39.636 | ||||||||
| T allele vs. C allele: OR =9.663, 95% CI: 1.312–71.149 | ||||||||
| Yan Q | BMC Cancer [2014] | China | PubMed, Web of Science | 2013.9.20 | Oral cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| TT vs. CC: OR =2.01, 95% CI: 0.75–5.41 | ||||||||
| CT vs. CC: OR =0.90, 95% CI: 0.55–1.47 | ||||||||
| TT + CT vs. CC: OR =1.04, 95% CI: 0.66–1.64 | ||||||||
| TT vs. CT + CC: OR =22.82, 95% CI: 0.28–1,887.72 | ||||||||
| T allele vs. C allele: OR =2.52, 95% CI: 0.71–8.98 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 4 | Risk: | ||||||
| AA vs. GG: OR =72.11, 95% CI: 2.08–2,502.44 | ||||||||
| GA vs. GG: OR =3.17, 95% CI: 1.26–7.92 | ||||||||
| AA + GA vs. GG: OR =7.92, 95% CI: 1.58–39.64 | ||||||||
| AA vs. GA + GG: OR =58.05, 95% CI: 1.70–1,985.77 | ||||||||
| A allele vs. G allele: OR =9.66, 95% CI: 1.31–71.15 | ||||||||
| Yang X | PLoS One [2013] | China | PubMed, Embase | 2013.6.26 | Oral cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| TT vs. CC: OR =2.01, 95% CI: 0.75–5.41 | ||||||||
| CT vs. CC: OR =0.85, 95% CI: 0.24–2.97 | ||||||||
| TT + CT vs. CC: OR =1.04, 95% CI: 0.61–1.78 | ||||||||
| TT vs. CT + CC: OR =22.8, 95% CI: 0.28–1,888 | ||||||||
| T allele vs. C allele: OR =3.93, 95% CI: 0.61–25.4 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| AA vs. GG: OR =20.7, 95% CI: 0.10–4519 | ||||||||
| GA vs. GG: OR =2.21, 95% CI: 0.18–26.9 | ||||||||
| AA + GA vs. GG: OR =7.81, 95% CI: 0.27–224 | ||||||||
| AA vs. GA + GG: OR =17.5, 95% CI: 0.10–3,257 | ||||||||
| A allele vs. G allele: OR =9.34, 95% CI: 0.23–388 | ||||||||
| Yang X | Tumour Biol [2014] | China | PubMed, Medline, Embase | 2013.7 | Oral cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk; |
| Homozygote codominant: OR =2.01, 95% CI: 0.75–5.41 | ||||||||
| Heterozygote codominant: OR =0.85, 95% CI: 0.24–2.97 | ||||||||
| Dominant model: OR =1.04, 95% CI: 0.61–1.78 | ||||||||
| Recessive model: OR =22.8, 95% CI: 0.28–1,887 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| Homozygote codominant: OR =20.7, 95% CI: 0.10–4519 | ||||||||
| Heterozygote codominant: OR =2.21, 95% CI: 0.18–26.9 | ||||||||
| Dominant model: OR =7.81, 95% CI: 0.27–225 | ||||||||
| Recessive model: OR =17.6, 95% CI: 0.10–3,257 | ||||||||
| Ye Y | Cancer Invest [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Oral carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| TT + CT vs. CC: OR =1.04, 95% CI: 0.60–1.80, P=0.9 | ||||||||
| Ye Y | Tumori [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Oral cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: |
| TT + CT vs. CC: OR =3.15, 95% CI: 1.05–9.47, P=0.04 |
Table S5
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Rainsbury JW | Head & Neck [2013] | UK | Cochrane, Medline, Zetoc, National Cancer Trials databases, Proquest Dissertations, Theses database, Conference Proceedings Citation Index | 2010.7 | Oropharyngeal squamous cell carcinoma | HIF-1α expression | 2 | OS: RR =1.27, 95% CI: 0.91–1.77 |
Table S6
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Jing S | Chinese Journal of Cancer Prevention and Treatment [2015]; Article in Chinese | China | PubMed, Embase, Cochrane, CBM, CNKI | 2014.1.30 | Nasopharyngeal carcinoma | HIF-1α expression | 6 | Risk: OR =0.052, 95% CI: 0.012–0.219, P<0.001 |
| 8 | Sex: OR =1.460, 95% CI: 0.939–2.268, P>0.05 | |||||||
| 6 | Age: OR =1.046, 95%CI: 0.389–2.812, P>0.05 | |||||||
| 5 | T1 + T2 vs. T3 + T4: OR =0.680, 95% CI: 0.423–1.092, P>0.05 | |||||||
| 7 | Lymph node metastasis: OR =0.296, 95% CI: 0.170–0.516, P<0.001 | |||||||
| 8 | Clinical stage: OR =0.298, 95% CI: 0.187–0.474, P<0.001 |
Table S7
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Anam MT | Biomark Res [2015] | Bangladesh | PubMed, PubMed Central, Google Scholar | 2014.12 | Lung cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: |
| TT vs. CC: OR =4.88, 95% CI: 2.42–9.84, P<0.0001 | ||||||||
| CT vs. CC: OR =1.56, 95% CI: 0.94–2.61, P=0.088 | ||||||||
| TT + CT vs. CC: OR =1.67, 95% CI: 0.79–3.54, P=0.1832 | ||||||||
| TT vs. CT + CC: OR =4.04, 95% CI: 2.02–8.08, P<0.0001 | ||||||||
| T allele vs. C allele: OR =1.68, 95% CI: 0.77–3.64, P=0.1908 | ||||||||
| Lung cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | |||||
| AA vs. GG: OR =5.41, 95% CI: 2.74–10.69, P<0.0001 | ||||||||
| GA vs. GG: OR =1.76, 95% CI: 1.25–2.49, P=0.0013 | ||||||||
| AA vs. GA + GG: OR =4.51, 95% CI: 2.31–8.81, P<0.0001 | ||||||||
| AA + GA vs. GG: OR =2.20, 95% CI: 1.60–3.03, P<0.0001 | ||||||||
| A allele vs. G allele: OR =2.31, 95% CI: 1.77–3.02, P<0.0001 | ||||||||
| He P | PLoS One [2013] | China | PubMed, Embase, CNKI | 2013.8.23 | Lung cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 3 | Dominant model (TT + CT vs. CC): OR = 1.19, 95% CI: 0.51–2.76 | |||||||
| 2 | Recessive model (TT vs. CT + CC): OR =1.39, 95% CI: 0.09–21.85 | |||||||
| 2 | Homozygote comparison (TT vs. CC): OR =1.42, 95% CI: 0.07–29.73 | |||||||
| 3 | Heterozygote comparison (CT vs. CC): OR =1.13, 95% CI: 0.59–2.19 | |||||||
| Hu X | Tumour Biol [2014] | China | PubMed, Embase, CNKI | 2013.7 | Lung cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| T allele vs. C allele: OR =1.19, 95% CI: 0.50–2.86 | ||||||||
| TT vs. CC: OR =1.41, 95% CI: 0.07–30.44 | ||||||||
| CT vs. CC: OR =1.13, 95% CI: 0.59–2.19 | ||||||||
| TT + CT vs. CC: OR =1.19, 95% CI: 0.51–2.76 | ||||||||
| TT vs. CT + CC: OR = 1.38, 95% CI: 0.09–22.18 | ||||||||
| Li C | Asian Pac J Cancer Prev [2013] | China | PubMed | 2012.12.20 | Non-small cell lung cancer | HIF-1α expression | 7 | OS: HR=1.50, 95% CI: 1.07–2.10 |
| HIF-2α expression | 3 | OS: HR=2.02, 95% CI: 1.47–2.77 | ||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Lung cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| TT vs. CC: OR =1.41, 95% CI: 0.07–30.44* | ||||||||
| CT vs. CC: OR =1.13, 95% CI: 0.59–2.19* | ||||||||
| TT/CT vs. CC: OR =1.19, 95% CI: 0.51–2.76* | ||||||||
| TT vs. CT/CC: OR =1.38, 95% CI: 0.09–22.18* | ||||||||
| T allele vs. C allele: OR =1.19, 95% CI: 0.50–2.86* | ||||||||
| Liao S | J Recept Signal Transduct Res [2015] | China | PubMed, Cochrane | 2014.9.1 | Lung cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: |
| CC vs. CT+TT: OR =0.50, 95% CI: 0.36–0.69, P<0.0001 | ||||||||
| TT vs. CT + CC: OR =4.04, 95% CI: 2.02–8.08, P<0.0001 | ||||||||
| T allele vs. C allele: OR =1.68, 95% CI: 0.77–3.64, P=0.19 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | ||||||
| GG vs. GA+AA: OR =0.45, 95% CI: 0.33–0.63, P<0.00001 | ||||||||
| AA vs. GA+GG: OR =4.52, 95% CI: 2.31–8.83, P<0.0001 | ||||||||
| A allele vs. G allele: OR =2.31, 95% CI: 1.77–3.02, P<0.00001 | ||||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Lung cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: |
| AA vs. GG: OR =5.42, 95% CI: 2.75–10.70, P<0.001 | ||||||||
| GA vs. GG: OR =1.72, 95% CI: 1.22–2.41, P=0.002 | ||||||||
| AA + GA vs. GG: OR =2.41, 95% CI: 1.56–2.94, P<0.001 | ||||||||
| AA vs. GA + GG: OR =4.52, 95% CI: 2.31–8.83, P<0.001 | ||||||||
| A allele vs. G allele: OR =2.26, 95% CI: 1.74–2.95, P<0.001 | ||||||||
| Ren W | Swiss Med Wkly [2013] | China | Cochrane, PubMed, Embase, CNKI, CBM, VIP, WanFang | 2012.5 | Lung cancer | HIF-1α expression | 4 | 5–year survival rates: OR = 0.13, 95% CI: 0.03–0.47, P=0.002 |
| 7 | OS: RR= 1.68, 95% CI: 1.12–2.50, P=0.01 | |||||||
| 16 | Tumor vs. benign tissues: OR =19.00, 95% CI: 12.12–29.78, P=0.00001 | |||||||
| 20 | Male vs. female: OR = 1.00, 95% CI: 0.80–1.26, P=0.99 | |||||||
| 12 | Age (≥60 vs. <60 years): OR = 1.14, 95% CI: 0.85–1.52, P=0.38 | |||||||
| 7 | Diameter (≥5 vs. <5 cm): OR = 1.84, 95% CI: 1.00–3.39, P=0.05 | |||||||
| 4 | Smoking vs. no smoking: OR = 2.16, 95% CI: 0.77–6.05, P=0.14 | |||||||
| 18 | Adenocarcinomas vs. squamous cell carcinoma: OR = 0.78, 95% CI: 0.63–0.98, P=0.03 | |||||||
| 4 | Non-small cell lung cancer vs. small cell lung cancer: OR = 0.24, 95% CI: 0.07–0.77, P=0.02 | |||||||
| 21 | Stage (I–II vs. III–IV): OR = 0.23, 95% CI: 0.14–0.36, P=0.00001 | |||||||
| 22 | Lymph node metastasis (yes vs. no): OR = 3.72, 95% CI: 2.38–5.80, P=0.00001 | |||||||
| 18 | Differentiation (well vs. poorly): OR = 0.47, 95% CI: 0.31–0.70, P=0.0002 | |||||||
| Wang Q | Gene [2014] | China | PubMed, Embase, Web of Science | 2013.8.31 | Non-small cell lung cancer | HIF-1α expression | 13 | OS: HR=1.60, 95% CI: 1.14–2.25, P=0.007 |
| Yan Q | BMC Cancer [2014] | China | PubMed, Web of Science | 2013.9.20 | Lung cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: |
| TT vs. CC: OR = 1.41, 95% CI: 0.07–30.44 | ||||||||
| CT vs. CC: OR =1.13, 95% CI: 0.59–2.19 | ||||||||
| TT + CT vs. CC: OR =1.19, 95% CI: 0.51–2.76 | ||||||||
| TT vs. CT + CC: OR =3.27, 95% CI: 1.73–6.17 | ||||||||
| T allele vs. C allele: OR =1.19, 95% CI: 0.50–2.86 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| AA vs. GG: OR =5.42, 95% CI: 2.74–10.70 | ||||||||
| GA vs. GG: OR =1.72, 95% CI: 1.22–2.41 | ||||||||
| AA + GA vs. GG: OR =2.14, 95% CI: 1.56–2.94 | ||||||||
| AA vs. GA + GG: OR =4.52, 95% CI: 2.31–8.83 | ||||||||
| A allele vs. G allele: OR =2.27, 95% CI: 1.74–2.95 | ||||||||
| Yang X | PLoS One [2013] | China | PubMed, Embase | 2013.6.26 | Lung cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: |
| TT vs. CC: OR =1.41, 95% CI: 0.07–30.4 | ||||||||
| CT vs. CC: OR =1.13, 95% CI: 0.59–2.19 | ||||||||
| TT + CT vs. CC: OR =1.50, 95% CI: 1.15–1.96 | ||||||||
| TT vs. CT + CC: OR =3.27, 95% CI: 1.73–6.17 | ||||||||
| T allele vs. C allele: OR =1.19, 95% CI: 0.50–2.86 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| AA vs. GG: OR =5.42, 95% CI: 2.75–10.7 | ||||||||
| GA vs. GG: OR =0.26, 95% CI: 0.01–7.10 | ||||||||
| AA + GA vs. GG: OR =0.82, 95% CI: 0.56–1.19 | ||||||||
| AA vs. GA + GG: OR =7.11, 95% CI: 3.61–14.0 | ||||||||
| A allele vs. G allele: OR =1.48, 95% CI: 1.09–2.00 | ||||||||
| Zhou Y | Cancer Cell Int [2014] | China | PubMed, Embase, CNKI | 2013.12.13 | Lung cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |
| 3 | AA + GA vs. GG: OR =2.14, 95% CI: 1.56–2.95 | |||||||
| 2 | AA vs. GA + GG: OR =4.5, 95% CI: 2.3–8.81 | |||||||
| 2 | AA vs. GG: OR =5.42, 95% CI: 2.74–10.7 | |||||||
| 2 | GA vs. GG: OR =3.02, 95% CI: 1.48–6.16 |
Notes: *In the study by Li Y, based on the 95% CI of OR, the statistical difference should not be significant.
Table S8
| First author | Journal (year) | No. studies | Included studies | No. Case | No. Control | Results | Model |
| He P | PLoS One [2013] | 3 | Kuo WH, et al. Transl Res [2012] | 285 | 300 | TT vs. CT + CC: OR =1.39, 95% CI: 0.09–21.85; P value for heterogeneity =0.07 | A fixed-effect model was used when P heterogeneity <0.05, otherwise a random effect model was used |
| Putra AC, et al. Respirology [2011] | 83 | 110 | |||||
| Konac E, et al. Exp Biol Med (Maywood) [2009] | 141 | 156 | |||||
| Hu X | Tumour Biol [2014] | 3 | Kuo WH, et al. Transl Res [2012] | 285 | 300 | TT vs. CT + CC: OR =1.38, 95% CI: 0.09–22.18; P value for heterogeneity =0.065 | A P value of more than 0.05 for the Q test indicated a lack of heterogeneity, and the fixed-effects model (the Mantel-Haenszel method) was subsequently used to calculate the summary ORs. Otherwise, the random-effects model (the DerSimonian and Laird method) was applied |
| Putra AC, et al. Respirology [2011] | 83 | 110 | |||||
| Konac E, et al. Exp Biol Med (Maywood) [2009] | 141 | 156 | |||||
| Li Y | Int J Clin Exp Med [2015] | 3 | Kuo WH, et al. Transl Res [2012] | 285 | 300 | TT vs. CT/CC: OR =1.38, 95% CI: 0.09-22.18; P value for heterogeneity =0.065 | Fixed effects model was used to pool the data when the P value of Q-test ≥0.05; otherwise, random effects model was selected |
| Putra AC, et al. Respirology [2011] | 83 | 110 | |||||
| Konac E, et al. Exp Biol Med (Maywood) [2009] | 141 | 156 | |||||
| Yan Q | BMC Cancer [2014] | 3 | Kuo WH, et al. Transl Res [2012] | 285 | 300 | TT vs. CT + CC: OR =3.27, 95% CI: 1.73–6.17; P value for heterogeneity =0.07 | When P > 0.05, the effects were assumed to be homogenous, and the fixed-effect model (the Mantel-Haenszel method) was used. When P<0.05, the random-effect model (the DerSimonian and Laird method) was more appropriate |
| Putra AC, et al. Respirology [2011] | 83 | 110 | |||||
| Konac E, et al. Exp Biol Med (Maywood) [2009] | 141 | 156 | |||||
| Yang X | PLoS One [2013] | 3 | Kuo WH, et al. Transl Res [2012] | 285 | 300 | TT vs. CT + CC: OR =3.27, 95% CI: 1.73–6.17; P value for heterogeneity =0.065 | A random-effects model was used when the significant Q statistic (P<0.1) indicated the presence of heterogeneity in the studies. Otherwise, a fixed-effects model was selected |
| Putra AC, et al. Respirology [2011] | 83 | 110 | |||||
| Konac E, et al. Exp Biol Med (Maywood) [2009] | 141 | 156 |
Table S9
| First author | Journal (year) | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Anam MT | Biomark Res [2015] | Bangladesh | PubMed, PubMed Central, Google Scholar | 2014.12 | Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: |
| TT vs. CC: OR =5.18, 95% CI: 0.88–30.38, P=0.0683 | ||||||||
| CT vs. CC: OR =1.00, 95% CI: 0.77–1.29, P=0.9964 | ||||||||
| TT + CT vs. CC: OR =1.05, 95% CI: 0.81–1.35, P=0.7221 | ||||||||
| TT vs. CT + CC: OR =5.18, 95% CI: 0.88–30.36, P=0.0684 | ||||||||
| T allele vs. C allele: OR =1.09, 95% CI: 0.86–1.39, P=0.4701 | ||||||||
| Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | |||||
| AA vs. GG: OR =0.36, 95% CI: 0.01–8.95, P=0.5332 | ||||||||
| GA vs. GG: OR =0.35, 95% CI: 0.10–1.24, P=0.1045 | ||||||||
| AA vs. GA + GG: OR =0.37, 95% CI: 0.02–9.29, P=0.5484 | ||||||||
| AA + GA vs. GG: OR =0.32, 95% CI: 0.09–1.10, P=0.0702 | ||||||||
| A allele vs. G allele: OR =0.30, 95% CI: 0.09–1.00, P=0.0495 | ||||||||
| He P | PLoS One [2013] | China | PubMed, Embase, CNKI | 2013.8.23 | Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |
| 6 | Dominant model (TT + CT vs. CC): OR =1.12, 95% CI: 0.87–1.52 | |||||||
| 5 | Recessive model (TT vs. CT + CC): OR =1.64, 95% CI: 0.56–4.77 | |||||||
| 5 | Homozygote comparison (TT vs. CC): OR =1.69, 95% CI: 0.56–5.14 | |||||||
| 6 | Heterozygote comparison (CT vs. CC): OR =1.10, 95% CI: 0.83–1.46 | |||||||
| Hu X | Tumour Biol [2013] | China | PubMed, Embase, CNKI | 2013.2 | Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 4 | Lymph node metastasis: OR =1.31, 95% CI: 0.98–1.75, P=0.069 |
| 3 | Histological grade: | |||||||
| Grades G3 vs. G1: OR =1.41, 95% CI: 0.70–2.85, P=0.336 | ||||||||
| Grades G3 vs. G2: OR =1.42, 95% CI: 0.91–2.20, P=0.121 | ||||||||
| Grades G2 vs. G1: OR =1.12, 95% CI: 0.56–2.24, P=0.745 | ||||||||
| Hu X | Tumour Biol [2014] | China | PubMed, Embase, CNKI | 2013.7 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 5 | Risk: |
| T allele vs. C allele: OR =1.09, 95% CI: 0.76–1.55 | ||||||||
| TT vs. CC: OR =2.16, 95% CI: 0.52–8.85 | ||||||||
| CT vs. CC: OR =1.05, 95% CI: 0.79–1.39 | ||||||||
| TT + CT vs. CC: OR =1.07, 95% CI: 0.76–1.50 | ||||||||
| TT vs. CT + CC: OR =2.15, 95% CI: 0.57–8.01 | ||||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 5 | Risk: |
| TT vs. CC: OR =2.16, 95% CI: 0.52–8.85, P=0.031 | ||||||||
| CT vs. CC: OR =1.07, 95% CI: 0.88–1.29, P=0.516 | ||||||||
| TT/CT vs. CC: OR =1.07, 95% CI: 0.76–1.50, P=0.254 | ||||||||
| TT vs. CT/CC: OR =2.27, 95% CI: 1.06–4.87, P=0.035 | ||||||||
| T allele vs. C allele: OR =1.09, 95% CI: 0.76–1.55, P=0.106 | ||||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: |
| AA vs. GG: OR =1.44, 95% CI: 0.38–5.44, P=0.595 | ||||||||
| GA vs. GG: OR =0.68, 95% CI: 0.23–2.05, P=0.498 | ||||||||
| AA + GA vs. GG: OR =0.63, 95% CI: 0.19–2.10, P=0.451 | ||||||||
| AA vs. GA + GG: OR =1.41, 95% CI: 0.37–5.37, P=0.613 | ||||||||
| A allele vs. G allele: OR =0.59, 95% CI: 0.17–2.10, P=0.419 | ||||||||
| Ren HT | Med Sci Monit [2014] | China | PubMed | 2013.6 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 6 | Risk: |
| TT vs. CC: OR =1.64, 95% CI: 0.85–3.19, P=0.14 | ||||||||
| CT vs. CC: OR =1.05, 95% CI: 0.87–1.27, P=0.58 | ||||||||
| TT + CT vs. CC: OR =1.13, 95% CI: 0.94–1.36, P=0.19 | ||||||||
| TT vs. CT + CC: OR =1.62, 95% CI: 0.83–3.15, P=0.16 | ||||||||
| T allele vs. C allele: OR =1.10, 95% CI: 0.93–1.30, P=0.28 | ||||||||
| Sun G | Breast J [2014] | China | NA | 2009 | Breast cancer | HIF-1α protein expression | 12 | Cancer vs. normal tissues: OR =23.11, 95% CI: 10.07–53.03, P<0.05 |
| 12 | Pathological differentiation: OR =3.77, 95% CI: 2.78–5.11, P<0.05 | |||||||
| 7 | Regional invasive extension (T3–4 vs. T1–2): OR =1.21, 95% CI: 0.87–1.87, P>0.05 | |||||||
| 10 | Axillary lymph node status (positive vs. negative): OR =3.03, 95% CI: 1.76–5.22, P<0.05 | |||||||
| 9 | Clinical stage: OR =2.82, 95% CI: 1.94–4.10, P<0.05 | |||||||
| 7 | VEGF expression: OR =1.21, 95% CI: 0.87–1.87, P<0.05 | |||||||
| 4 | Overall survival: OR =0.54, 95% CI: 0.35–0.83, P<0.05 | |||||||
| Wang W | Clinica Chimica Acta [2014] | China | PubMed, Embase, Web of Science | 2013.4.1 | Breast cancer | HIF-1α expression | 7 | OS: HR=1.46, 95% CI: 1.12–1.92, P=0.006 |
| 8 | DFS: HR=1.91, 95% CI: 1.43–2.57, P<0.001 | |||||||
| 3 | DMFS: HR=2.17, 95% CI: 1.16–4.05, P=0.015 | |||||||
| 3 | RFS: HR=1.33, 95% CI: 1.09–1.61, P=0.005 | |||||||
| Wu G | Tumour Biol [2014] | China | PubMed, Embase, Google Scholar, Wanfang | 2013.6.10 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 6 | TT + CT vs. CC: OR =0.99, 95% CI: 0.72–1.36, P=0.951 | |||||||
| 6 | TT vs. CT + CC: OR =1.05, 95% CI: 0.88–1.25, P=0.561 | |||||||
| Yan Q | BMC Cancer [2014] | China | PubMed, Web of Science | 2013.9.20 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 6 | Risk: |
| TT vs. CC: OR =1.41, 95% CI: 0.34–5.75 | ||||||||
| CT vs. CC: OR =1.01, 95% CI: 0.91–1.33 | ||||||||
| TT + CT vs. CC: OR =1.13, 95% CI: 0.94–1.36 | ||||||||
| TT vs. CT + CC: OR =1.38, 95% CI: 0.35–5.46 | ||||||||
| T allele vs. C allele: OR =1.09, 95% CI: 0.80–1.48 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 4 | Risk: | ||||||
| AA vs. GG: OR =1.44, 95% CI: 0.38–5.44 | ||||||||
| GA vs. GG: OR =1.03, 95% CI: 0.70–1.52 | ||||||||
| AA + GA vs. GG: OR =1.05, 95% CI: 0.72–1.53 | ||||||||
| AA vs. GA + GG: OR =1.41, 95% CI: 0.37–5.40 | ||||||||
| A allele vs. G allele: OR =1.07, 95% CI: 0.76–1.52 | ||||||||
| Yang X | PLoS One [2013] | China | PubMed, Embase | 2013.6.26 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 5 | Risk: |
| TT vs. CC: OR =2.30, 95% CI: 1.08–4.91 | ||||||||
| CT vs. CC: OR =1.07, 95% CI: 0.88–1.29 | ||||||||
| TT + CT vs. CC: OR =1.12, 95% CI: 0.92–1.35 | ||||||||
| TT vs. CT + CC: OR =2.27, 95% CI: 1.06–4.87 | ||||||||
| T allele vs. C allele: OR =1.09, 95% CI: 0.76–1.55 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| AA vs. GG: OR =1.44, 95% CI: 0.38–5.44 | ||||||||
| GA vs. GG: OR =1.03, 95% CI: 0.70–1.52 | ||||||||
| AA + GA vs. GG: OR =1.05, 95% CI: 0.72–1.53 | ||||||||
| AA vs. GA + GG: OR =1.41, 95% CI: 0.37–5.37 | ||||||||
| A allele vs. G allele: OR =1.07, 95% CI: 0.75–1.52 | ||||||||
| Ye Y | Cancer Invest [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: TT + CT vs. CC: OR =0.91, 95% CI: 0.62–1.32, P=0.51 |
| Ye Y | Tumori [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: TT + CT vs. CC: OR =0.32, 95% CI: 0.09–1.10, P=0.07 |
| Ye Y | Tumori [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: TT + CT vs. CC: OR =0.32, 95% CI: 0.09–1.10, P=0.07 |
| Yin W | Cancer Res (abstract) [2011] | China | NA | NA | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | NA | Risk: |
| Recessive model: OR =2.273, 95% CI: 1.061–4.872, P=0.035 | ||||||||
| Dominant model: OR =1.075, 95% CI: 0.717–1.613, P=0.725 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | NA | Risk: | ||||||
| Recessive model: not significant | ||||||||
| Dominant model: not significant | ||||||||
| Zhao T | J Exp Clin Cancer Res [2009] | China | PubMed | 2009.6 | Breast cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| T allele vs. C allele: OR =0.99, 95% CI: 0.79–1.23, P=0.9 | ||||||||
| TT vs. CT + CC: OR =1.51, 95% CI: 0.55–4.11, P=0.42 | ||||||||
| TT + CT vs. CC: OR =0.96, 95% CI: 0.76–1.21, P=0.75 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | ||||||
| A allele vs. G allele: OR =0.28, 95% CI: 0.08–0.90, P=0.03 | ||||||||
| AA + GA vs. GG: OR =0.29, 95% CI: 0.09–0.97, P=0.04 | ||||||||
| Zhou Y | Cancer Cell Int [2014] | China | PubMed, Embase, CNKI | 2013.12.13 | Breast cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |
| 3 | AA + GA vs. GG: OR =0.63, 95% CI: 0.19–2.08 | |||||||
| 2 | AA vs. GA + GG: OR =1.44, 95% CI: 0.34–6.08 | |||||||
| 2 | AA vs. GG: OR =1.43, 95% CI: 0.37–5.44 | |||||||
| 2 | GA vs. GG: OR =1.45, 95% CI: 0.34–6.17 |
Table S10
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Anam MT | Biomark Res [2015] | Bangladesh | PubMed, PubMed Central, Google Scholar | 2014.12 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| TT vs. CC: OR =1.91, 95% CI: 0.32–11.58, P=0.4801 | ||||||||
| CT vs. CC: OR =0.83, 95% CI: 0.50–1.39, P=0.4817 | ||||||||
| TT + CT vs. CC: OR =1.24, 95% CI: 0.77–2.01, P=0.3756 | ||||||||
| TT vs. CT + CC: OR =1.97, 95% CI: 0.33–11.90, P=0.4603 | ||||||||
| T allele vs. C allele: OR =0.94, 95% CI: 0.59–1.49, P=0.7833 | ||||||||
| Cao S | Clin Res Hepatol Gastroenterol [2014] | China | PubMed, Embase | 2013.8 | Hepatocellular carcinoma | HIF-1α protein expression | 4 | DFS: OR =2.10, 95% CI: 1.48–2.97 |
| 3 | Capsule formation: OR =1.25, 95% CI: 0.93–1.69 | |||||||
| 4 | Cirrhosis: OR =1.00, 95% CI: 0.76–1.30 | |||||||
| 6 | Tumor size: OR =0.92, 95% CI: 0.74–1.14 | |||||||
| 3 | Tumor differentiation: OR =0.89, 95% CI: 0.65–1.21 | |||||||
| 4 | Vascular invasion: OR =2.04, 95% CI: 1.31–3.18 | |||||||
| 5 | HCC tissue vs. paraneoplastic tissue: OR =2.50, 95% CI: 0.98–6.36 | |||||||
| Chen J | PLoS One [2014] | China | PubMed, Embase, Cochrane, CNKI | 2013.6 | Gastric cancer | HIF-1α protein expression | 10 | 5–year OS: RR=1.508, 95% CI: 1.318–1.725, P<0.001 |
| 9 | Depth of invasion (T3 and T4 vs. T1 and T2): OR =3.050, 95% CI: 2.067–4.501, P<0.001 | |||||||
| 11 | Lymph node status: OR =3.486, 95% CI: 2.737–4.440, P<0.001 | |||||||
| 5 | Distant metastasis: OR =6.635, 95% CI: 1.855–23.738, P=0.004 | |||||||
| 10 | TNM stage (stages III and IV vs. stage I and II): OR =2.762, 95% CI: 1.941–3.942, P<0.001 | |||||||
| 6 | Vascular invasion: OR =2.368, 95% CI: 1.725–3.252, P<0.001 | |||||||
| 10 | Histological differentiation: OR =2.112, 95% CI: 1.410–3.163, P<0.001 | |||||||
| 5 | Size: OR =1.921, 95% CI: 1.395–2.647, P<0.001 | |||||||
| 7 | Sex: OR =0.905, 95% CI: 0.679–1.205, P=0.495 | |||||||
| 11 | Age: OR =0.846, 95% CI: 0.667–1.072, P=0.166 | |||||||
| Chen Z | PLoS One [2013] | China | PubMed, Wanfang, Web of Science | NA | Colorectal cancer | HIF-1α protein expression | 9 | DFS: HR=2.84, 95% CI: 1.87–4.31 |
| 11 | OS: HR=2.01, 95% CI: 1.55–2.6 | |||||||
| 15 | Differentiation grade: OR =0.97, 95% CI: 0.67–1.39, P=0.864 | |||||||
| 5 | Dukes’ stages: OR =0.39, 95% CI: 0.17–0.89, P=0.025 | |||||||
| 15 | Lymph node status: OR =0.49, 95% CI: 0.32–0.73, P=0.001 | |||||||
| 9 | Depth of invasion: OR =0.71, 95% CI: 0.51–0.99, P=0.045 | |||||||
| 5 | Metastasis: OR =0.29, 95% CI: 0.11–0.81, P=0.018 | |||||||
| 9 | UICC stage: OR =0.42, 95% CI: 0.3–0.59, P<0.001 | |||||||
| HIF-2α protein expression | 4 | OS: HR=2.07, 95% CI: 1.01–4.26 | ||||||
| 2 | Differentiation grade: OR =0.484, 95% CI: 0.289–0.812, P=0.006 | |||||||
| 2 | Dukes’ stages: OR =0.9, 95% CI: 0.197–4.168, P=0.9 | |||||||
| 3 | Lymph node status: OR =0.95, 95% CI: 0.418–2.16, P=0.904 | |||||||
| 2 | Depth of invasion: OR =0.379, 95% CI: 0.038–3.798, P=0.409 | |||||||
| He P | PLoS One [2013] | China | PubMed, Embase, CNKI | 2013.8.23 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 2 | Dominant model (TT + CT vs. CC): OR =0.26, 95% CI: 0.01–5.09 | |||||||
| 1 | Recessive model (TT vs. CT + CC): OR =1.97, 95% CI: 0.33–11.90 | |||||||
| 1 | Homozygote comparison (TT vs. CC): OR =1.91, 95% CI: 0.32–11.58 | |||||||
| 2 | Heterozygote comparison (CT vs. CC): OR =0.25, 95% CI: 0.01–4.69 | |||||||
| Pancreatic cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | ||||||
| 2 | Dominant model (TT + CT vs. CC): OR = 1.39, 95% CI: 0.54 –3.56 | |||||||
| 1 | Recessive model (TT vs. CT + CC): OR =4.13, 95% CI: 1.57–10.86 | |||||||
| 1 | Homozygote comparison (TT vs. CC): OR =3.39, 95% CI: 1.28–8.97 | |||||||
| 2 | Heterozygote comparison (CT vs. CC): OR =0.51, 95% CI: 0.02–11.53 | |||||||
| Hu X | Tumour Biol [2013] | China | PubMed, Embase, CNKI | 2013.2 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Lymph node metastasis: OR =1.23, 95% CI: 0.73–2.07, P=0.429 |
| 2 | Histological grade: | |||||||
| Grades G3 vs. G1: OR =0.58, 95% CI: 0.13–2.53, P=0.47 | ||||||||
| Grades G3 vs. G2: OR =1.24, 95% CI: 0.32–4.89, P=0.757 | ||||||||
| Grades G2 vs. G1: OR =0.52, 95% CI: 0.25–1.10, P=0.086 | ||||||||
| Hu X | Tumour Biol [2014] | China | PubMed, Embase, CNKI | 2013.7 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| T allele vs. C allele: OR =0.26, 95% CI: 0.01–6.38 | ||||||||
| TT vs. CC: OR =1.91, 95% CI: 0.32–11.58 | ||||||||
| CT vs. CC: OR =0.24, 95% CI: 0.01–5.51 | ||||||||
| TT + CT vs. CC: OR =1.17, 95% CI: 0.62–2.22 | ||||||||
| TT vs. CT + CC: OR =1.97, 95% CI: 0.33–11.90 | ||||||||
| Jing S | Chin J Pathol [2014]Article in Chinese | China | PubMed, Embase, Cochrane, CBM, CNKI | 2014.7.30 | Esophageal squamous cell carcinoma | HIF-1α protein expression | 8 | Risk: OR =0.088, 95% CI: 0.061–0.129, P<0.001 |
| 10 | Tumor differentiation: OR =1.287, 95% CI: 0.904–1.831, P=0.161 | |||||||
| 4 | Histological grade: OR =1.194, 95% CI: 0.307–4.642, P=0.798 | |||||||
| 8 | T1 + T2 vs. T3 + T4: OR =0.421, 95% CI: 0.222–0.798, P=0.008 | |||||||
| 14 | Lymph node metastasis: OR =0.387, 95% CI: 0.207–0.725, P=0.003 | |||||||
| 8 | Tumor stage: OR =0.525, 95% CI: 0.236–1.171, P=0.116 | |||||||
| 5 | Lymphatic vessels invasion: OR =0.560, 95% CI: 0.219–1.431, P=0.226 | |||||||
| 5 | Vascular invasion: OR =0.971, 95% CI: 0.667–1.413, P=0.877 | |||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| TT vs. CC: OR =1.91, 95% CI: 0.32–11.58 | ||||||||
| CT vs. CC: OR =0.34, 95% CI: 0.09–1.34* | ||||||||
| TT/CT vs. CC: OR =0.34, 95% CI: 0.08–1.41* | ||||||||
| TT vs. CT/CC: OR =1.97, 95% CI: 0.33–11.90* | ||||||||
| T allele vs. C allele: OR =0.38, 95% CI: 0.09–1.50* | ||||||||
| Esophageal squamous cell carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: | |||||
| CT vs. CC: OR =1.11, 95% CI: 0.46–2.69, P=0.822 | ||||||||
| TT/CT vs. CC: OR =1.11, 95% CI: 0.46–2.69, P=0.822 | ||||||||
| T allele vs. C allele: OR =1.10, 95% CI: 0.47–2.60, P=0.827 | ||||||||
| Pancreatic cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: | |||||
| CT vs. CC: OR =2.16, 95% CI: 1.32–3.51, P=0.002TT/CT vs. CC: OR =2.16, 95% CI: 1.32–3.51, P=0.002 | ||||||||
| T allele vs. C allele: OR =2.02, 95% CI: 1.27–3.23, P=0.003 | ||||||||
| Hepatocellular carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: | |||||
| CT vs. CC: OR =2.19, 95% CI: 0.88–5.43, P=0.092 | ||||||||
| TT/CT vs. CC: OR =2.19, 95% CI: 0.88–5.43, P=0.092 | ||||||||
| T allele vs. C allele: OR =2.14, 95% CI: 0.87–5.23, P=0.096 | ||||||||
| Gastric cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: | |||||
| CT vs. CC: OR =0.34, 95% CI: 0.11–1.10, P=0.072 | ||||||||
| TT/CT vs. CC: OR =0.34, 95% CI: 0.11–1.10, P=0.072 | ||||||||
| T allele vs. C allele: OR =0.36, 95% CI: 0.12–1.13, P=0.079 | ||||||||
| Lin S | World J Gastroenterol [2014] | China | PubMed, Embase, Web of Science | 2013.8 | Gastric cancer | HIF-1α expression | 5 | 5-year OS rate: OR =0.36, 95% CI: 0.21–0.64, P=0.0004 |
| 6 | Tumor differentiation: OR =0.38, 95% CI: 0.23–0.64, P=0.0003 | |||||||
| 7 | Depth of invasion: OR =0.42, 95% CI: 0.32–0.57, P<0.00001 | |||||||
| 9 | Lymph node metastasis: OR =2.23, 95% CI: 1.46–3.40, P=0.0002 | |||||||
| 5 | Lymphatic invasion: OR =2.50, 95% CI: 1.46–4.28, P=0.0009 | |||||||
| 5 | Vascular invasion: OR =1.80, 95% CI: 1.29–2.51, P=0.0005 | |||||||
| 6 | TNM stages III + IV: OR =0.31; 95% CI: 0.15–0.60, P=0.0006 | |||||||
| Liu J | Gene [2013] | China | PubMed, Embase | 2012.3 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | NA | Risk: OR =1.239, 95% CI =0.985–1.559, P=0.067 |
| HIF-1α rs11549467 (1790 G/A) polymorphism | NA | Risk: OR =0.867, 95% CI =0.492–1.528, P=0.622 | ||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Colorectal cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: |
| GA vs. GG: OR =0.97, 95% CI: 0.57–1.63, P=0.912 | ||||||||
| AA + GA vs. GG: OR =0.97, 95% CI: 0.57–1.63, P=0.912 | ||||||||
| A allele vs. G allele: OR =0.97, 95% CI: 0.58–1.62, P=0.914 | ||||||||
| Pancreatic cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | |||||
| AA vs. GG: OR =9.30, 95% CI: 1.12–77.61, P=0.039 | ||||||||
| GA vs. GG: OR =2.90, 95% CI: 1.82–4.62, P=0.625 | ||||||||
| AA + GA vs. GG: OR =2.50, 95% CI: 0.93–6.73, P=0.070 | ||||||||
| AA vs. GA + GG: OR =8.65, 95% CI: 1.04–71.65, P=0.045 | ||||||||
| A allele vs. G allele: OR =3.12, 95% CI: 2.01–4.84, P<0.001 | ||||||||
| Hepatocellular carcinoma | HIF-1α rs11549467 (1790 G/A) polymorphism | 1 | Risk: | |||||
| GA vs. GG: OR =4.10, 95% CI: 1.91–8.82, P<0.001 | ||||||||
| AA + GA vs. GG: OR =4.10, 95% CI: 1.91–8.82, P=0.006 | ||||||||
| A allele vs. G allele: OR =3.85, 95% CI:1.83–8.13, P<0.001 | ||||||||
| Gastric cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 1 | Risk: | |||||
| GA vs. GG: OR =2.93, 95% CI: 1.06–8.06, P=0.038 | ||||||||
| AA + GA vs. GG: OR =2.93, 95% CI: 1.06–8.06, P=0.038 | ||||||||
| A allele vs. G allele: OR =2.77, 95% CI:1.03–7.45, P=0.043 | ||||||||
| Ni Z | Genes Genom [2015] | China | NA | NA | Overall digestive tract cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 10 | Risk: |
| Allele model: OR =1.292, 95% CI =1.107–1.507, P=0.001 | ||||||||
| Dominant model: OR =1.277, 95% CI =1.083–1.507, P=0.004 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 9 | Risk: | ||||||
| Allele model: OR =1.920, 95% CI = 1.213–3.038, P=0.005 | ||||||||
| Dominant model: OR =1.957, 95% CI =1.219–3.142, P=0.005 | ||||||||
| Ping W | Tumour Biol [2014] | China | PubMed, Medline, Embase, Cochrane, Web of Science, CBM | 2013.9.10 | Esophageal squamous cell carcinoma | HIF-1α expression | 10 | OS: HR=1.84, 95% CI: 1.36–2.50, P<0.001 |
| 2 | DFS: HR= 2.00, 95% CI: 1.05–3.79, P=0.035 | |||||||
| 9 | Gender (male vs. female): HR= 0.82, 95% CI: 0.50–1.35, P=0.429 | |||||||
| 8 | Stage (stage III/IV vs. stage I/II): HR=2.90, 95% CI: 1.90–4.44, P<0.001 | |||||||
| 11 | Lymph node metastasis (yes vs. no): HR=1.93, 95% CI: 1.35–2.76, P<0.001 | |||||||
| 7 | Depth of invasion (T3/T4 vs. T1/T2): HR=2.45, 95% CI: 1.24–4.86, P=0.01 | |||||||
| 5 | Lymphatic invasion (yes vs. no): HR=2.25, 95% CI: 1.3–3.76, P=0.002 | |||||||
| 5 | Vascular invasion (yes vs. no): HR=1.34, 95% CI: 0.79–2.26, P=0.271 | |||||||
| 8 | Histological grade (poor vs. well/moderate): HR=1.20, 95% CI: 0.70–2.07, P=0.507 | |||||||
| 5 | Distant metastasis (M1 vs. M0): HR=1.97, 95% CI: 1.10–3.53, P=0.022 | |||||||
| 4 | Vascular endothelial growth factor (high vs. low): HR=3.67, 95% CI: 1.81–7.46, P<0.001 | |||||||
| Sun G | J Chin Oncol [2012] Article in Chinese | China | PubMed, Cochrane | 2011.12 | Esophageal squamous cell carcinoma | HIF-1α protein expression | 7 | Risk: OR =33.111, 95% CI: 11.912–92.040, P<0.001 |
| 11 | 2–year OS rate: RR=0.320, 95% CI: 0.115–0.887, P=0.0004 | |||||||
| 3 | Tumor differentiation: OR =1.185, 95% CI: 0.859–1.635, P=0.3 | |||||||
| 8 | Clinical stage: OR =0.421, 95% CI: 0.222–0.798, P=0.008 | |||||||
| 13 | Lymphoma node metastasis: OR =2.393, 95% CI: 1.319–4.344, P=0.003 | |||||||
| 9 | Depth of invasion: OR =1.701, 95% CI: 1.076–4.710, P=0.226 | |||||||
| Sun X | World J Gastroenterol [2015] | China | PubMed, Embase, CNKI | 2013.7.15 | Overall digestive tract cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 13 | Risk: |
| CT vs. CC: OR =0.853, 95% CI: 0.502–1.450 | ||||||||
| TT + CT vs. CC: OR =1.156, 95% CI: 0.839–1.593 | ||||||||
| T allele vs. C allele: OR =1.325, 95% CI: 0.846–2.076 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 10 | Risk: | ||||||
| GA vs. GG: OR =2.677, 95% CI: 1.677–4.273 | ||||||||
| AA + GA vs. GG: OR =3.252, 95% CI: 1.661–6.368 | ||||||||
| A allele vs. G allele: OR =4.455, 95% CI: 1.938–10.241 | ||||||||
| Pancreatic cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: | |||||
| CT vs. CC: OR =0.500, 95% CI: 0.018–14.015 | ||||||||
| TT + CT vs. CC: OR =1.388, 95% CI: 0.542–3.555 | ||||||||
| T allele vs. C allele: OR =1.753, 95% CI: 1.225–2.508 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | ||||||
| CT vs. CC: OR =1.611, 95% CI: 0.241–10.760 | ||||||||
| TT + CT vs. CC: OR =2.499, 95% CI: 0.929–6.726 | ||||||||
| T allele vs. C allele: OR =3.030, 95% CI: 1.946–4.716 | ||||||||
| Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: | |||||
| CT vs. CC: OR =0.241, 95% CI: 0.011–5.509 | ||||||||
| TT + CT vs. CC: OR =1.118, 95% CI: 0.573–2.182 | ||||||||
| T allele vs. C allele: OR =0.262, 95% CI: 0.011–6.380 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | ||||||
| TT + CT vs. CC: OR =0.971, 95% CI: 0.571–1.650 | ||||||||
| Wu G | Tumour Biol [2014] | China | PubMed, Embase, Google Scholar, Wanfang | 2013.6.10 | Overall digestive tract cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 9 | TT + CT vs. CC: OR =1.17, 95% CI: 0.78–1.75, P=0.441 | |||||||
| 7 | TT vs. CT + CC: OR =1.04, 95% CI: 0.63–1.71, P=0.879 | |||||||
| Xu J | Genet Mol Res [2014] | China | CISCOM, CINAHL, Web of Science, PubMed, Google Scholar, EBSCO, Cochrane, CBM | 2013.5.1 | Overall digestive tract cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 6 | Risk: |
| TT + CT vs. CC: OR =2.04, 95% CI: 1.06–3.92 | ||||||||
| T allele vs. C allele: OR =1.36, 95% CI: 1.15–1.62 | ||||||||
| Xu J | Genet Test Mol Biomarkers [2013] | China | PubMed, Embase, Web of Science, Cochrane, CBM | 2013.5.1 | Overall digestive tract cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 6 | Risk: |
| C allele vs. T allele: OR =1.36, 95% CI: 1.15–1.62, P<0.001 | ||||||||
| CC vs. TT + CT: OR =2.04, 95% CI: 1.06–3.92, P<0.001 | ||||||||
| Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: | |||||
| C allele vs. T allele: OR =0.27, 95% CI: 0.01–5.45, P=0.395 | ||||||||
| CC vs. TT + CT: OR =1.12, 95% CI: 0.58–2.17, P=0.738 | ||||||||
| Esophageal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: | |||||
| C allele vs. T allele: OR =1.10, 95% CI: 0.47–2.60, P=0.827 | ||||||||
| CC vs. TT + CT: OR =1.11, 95% CI: 0.46–2.69, P=0.822 | ||||||||
| Gastric cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: | |||||
| C allele vs. T allele: OR =5.17, 95% CI: 1.75–15.26, P=0.003 | ||||||||
| CC vs. TT+CT: OR =5.75, 95% CI: 1.91–17.35, P=0.002 | ||||||||
| Xu JJ | Genet Mol Res [2014] | China | PubMed, Embase, Web of Science, Cochrane, CBM | 2013.5.1 | Overall digestive tract cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 8 | Risk: |
| TT vs. CC: OR =1.91, 95% CI: 0.32–11.58, P=0.480 | ||||||||
| TT vs. CT: OR =2.30, 95% CI: 0.36–14.67, P=0.377 | ||||||||
| TT + CT vs. CC: OR =1.23, 95% CI: 0.79–1.91, P=0.367 | ||||||||
| TT vs. CT + CC: OR =1.97, 95% CI: 0.33–11.9, P=0.460 | ||||||||
| T allele vs. C allele: OR =1.03, 95% CI: 0.56–1.89, P=0.920 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 5 | Risk: | ||||||
| AA + GA vs. GG: OR =2.19, 95% CI: 1.12–4.29, P=0.022 | ||||||||
| A allele vs. G allele: OR =2.89, 95% CI: 1.91–4.37, P<0.001 | ||||||||
| Yan Q | BMC Cancer [2014] | China | PubMed, Web of Science | 2013.9.20 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| CT vs. CC: OR =0.24, 95% CI: 0.01–5.51 | ||||||||
| CT+ vs. CC: OR =1.12, 95% CI: 0.57–2.18 | ||||||||
| T allele vs. C allele: OR =0.26, 95% CI: 0.01–6.38 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: AA + GA vs. GG: OR =0.97, 95% CI: 0.57–1.63 | ||||||
| Pancreatic cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | |||||
| GA vs. GG: OR =1.61, 95% CI: 0.24–10.76 | ||||||||
| AA + GA vs. GG: OR =3.14, 95% CI: 1.99–4.97 | ||||||||
| A allele vs. G allele: OR =3.08, 95% CI: 1.98–4.78 | ||||||||
| Yang X | PLoS One [2013] | China | PubMed, Embase | 2013.6.26 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| TT vs. CC: OR =1.91, 95% CI: 0.32–11.6 | ||||||||
| CT vs. CC: OR =0.24, 95% CI: 0.01–5.51 | ||||||||
| TT + CT vs. CC: OR =1.10, 95% CI: 0.87–1.38 | ||||||||
| TT vs. CT + CC: OR =1.97, 95% CI: 0.33–11.9 | ||||||||
| T allele vs. C allele: OR =1.36, 95% CI: 0.68–2.70 | ||||||||
| Yang X | Tumour Biol [2014] | China | PubMed, Medline, Embase | 2013.7 | Overall digestive tract cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 12 | Risk: |
| Homozygote codominant: OR =2.51, 95% CI: 1.31–4.81 | ||||||||
| Heterozygote codominant: OR =0.81, 95% CI: 0.45–1.48 | ||||||||
| Dominant model: OR =1.16, 95% CI: 0.82–1.64 | ||||||||
| Recessive model: OR =8.73, 95% CI: 1.33–57.1 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 9 | Risk: | ||||||
| Homozygote codominant: OR =14.6, 95% CI: 0.70–305 | ||||||||
| Heterozygote codominant: OR =2.26, 95% CI: 0.91–5.59 | ||||||||
| Dominant model: OR =3.17, 95% CI: 1.21–8.25 | ||||||||
| Recessive model: OR =12.8, 95% CI: 0.65–252 | ||||||||
| Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: | |||||
| Homozygote codominant: OR =1.91, 95% CI: 0.32–11.6 | ||||||||
| Heterozygote codominant: OR =0.24, 95% CI: 0.01–5.51 | ||||||||
| Dominant model: OR =1.12, 95% CI: 0.57–2.18 | ||||||||
| Recessive model: OR =1.97, 95% CI: 0.33–11.9 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | ||||||
| Heterozygote codominant: OR =1.31, 95% CI: 0.51–3.36 | ||||||||
| Dominant model: OR =0.97, 95% CI: 0.57–1.63 | ||||||||
| Pancreatic cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: | |||||
| Homozygote codominant: OR =3.39, 95% CI: 1.28–8.97 | ||||||||
| Heterozygote codominant: OR =0.50, 95% CI: 0.02–14.0 | ||||||||
| Dominant model: OR =1.39, 95% CI: 0.54–3.56 | ||||||||
| Recessive model: OR =4.13, 95% CI: 1.57–10.9 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | ||||||
| Homozygote codominant: OR =9.30, 95% CI: 1.12–77.6 | ||||||||
| Heterozygote codominant: OR =1.60, 95% CI: 0.24–10.52 | ||||||||
| Dominant model: OR =3.14, 95% CI: 1.99–4.97 | ||||||||
| Recessive model: OR =8.65, 95% CI: 1.05–71.6 | ||||||||
| Yao Q | Saudi Med J [2015] | China | PubMed, Embase, Web of Science, Elsevier Science Direct, CBM, CNKI | 2014.2.28 | Hepatocellular carcinoma | HIF-2α expression | 5 | OS: HR=1.640, 95% CI: 0.648–4.151 |
| 5 | Tumor size: OR =2.173, 95% CI: 0.553–8.533, P=0.226 | |||||||
| 4 | Capsule infiltration: OR =2.738, 95% CI: 1.709–4.386, P<0.001 | |||||||
| 3 | Vein invasion: OR =2.458, 95% CI: 1.053–5.734, P=0.038 | |||||||
| 4 | Liver cirrhosis: OR =1.179, 95% CI: 0.525–2.647, P=0.690 | |||||||
| 5 | Histological grade: OR =0.172, 95% CI: 0.042–0.713, P=0.015 | |||||||
| 3 | Necrosis: OR =2.362, 95% CI: 0.472–11.815, P=0.295 | |||||||
| Ye LY | Pancreatology [2014] | China | Medline, Embase, Web of Science, Manual search | NA | Pancreatic cancer | HIF-1α expression | 6 | OS: HR=1.88, 95% CI: 1.39–2.56, P<0.05 |
| 6 | Lymph node metastasis: OR =3.16, 95% CI: 1.95–5.11, P<0.05 | |||||||
| 4 | Tumor size: OR =1.58, 95% CI: 0.46–5.47, p>0.05 | |||||||
| 3 | Tumor staging (I–II vs. III–IV): OR =3.66, 95% CI: 2.01–6.69, P<0.05 | |||||||
| Ye Y | Cancer Invest [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: TT + CT vs. CC: OR =0.88, 95% CI: 0.46–1.68, P=0.7 |
| Ye Y | Tumori [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Overall digestive tract cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 5 | Risk: TT + CT vs. CC: OR =2.20, 95% CI: 1.12–4.34, P=0.02 |
| Zhang ZG | Asian Pac J Cancer Prev [2013] | China | Cochrane, PubMed, EMBASE, Web of Science, CBM | 2013.2 | Gastric cancer | HIF-1α expression | 10 | OS: HR =1.34, 95% CI: 1.13–1.58, P=0.0009 |
| 5 | DFS: HR =1.67, 95% CI: 0.99–2.82, P=0.06 | |||||||
| Zhao T | J Exp Clin Cancer Res [2009] | China | PubMed | 2009.6 | Colorectal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: |
| T allele vs. C allele: OR =0.26, 95% CI: 0.01–6.38, P=0.41 | ||||||||
| TT vs. CT + CC: OR =1.97, 95% CI: 0.33–11.90, P=0.46 | ||||||||
| TT + CT vs. CC: OR =0.25, 95% CI: 0.01–5.99, P=0.39 | ||||||||
| Zheng F | Medicine [2016] | China | PubMed, Cochrane, EBSCO | NA | Gastric cancer | HIF-2α expression | 2 | 5-year OS rate: OR =2.08, 95% CI: 1.21–3.58, P=0.0008 |
| 4 | Tumor infiltration (T3 and T4 vs. T2 and T1): OR =3.08, 95% CI: 1.18–8.04, P=0.022 | |||||||
| 5 | Lymphatic metastasis: OR =3.26, 95% CI: 1.10–9.63, P=0.033 | |||||||
| 5 | TNM stage: OR =2.61, 95% CI: 1.40–4.84, P=0.002 | |||||||
| 3 | Tumor differentiation: OR =2.03, 95% CI: 0.73–5.64, P=0.173 | |||||||
| Zheng SS | PLoS One [2013] | China | PubMed, Elsevier, Web of Science | 2013.2 | Hepatocellular carcinoma | HIF-1α protein expression | 3 | DFS: HR=2.14, 95% CI: 1.39–3.29 |
| 6 | OS: HR=1.65, 95% CI: 1.38–1.97 | |||||||
| Zhou Y | Cancer Cell Int [2014] | China | PubMed, Embase, CNKI | 2013.12.13 | Pancreatic cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |
| 2 | AA + GA vs. GG: OR =2.5, 95% CI: 0.93–6.72 | |||||||
| 1 | AA vs. GA + GG: OR =18.8, 95% CI: 0.96–371.55 | |||||||
| 1 | AA vs. GG: OR =18.3, 95% CI: 0.93–360.19 | |||||||
| 1 | GA vs. GG: OR =29.4, 95% CI: 1.12–772.37 | |||||||
| Zhu C | Mol Biol Rep [2013] | China | PubMed, Embase, Web of Science | 2012.12.1 | Gastric cancer | HIF-1α expression | 9 | Overall mortality risk: HR =2.14, 95% CI: 1.32–3.48 |
| 4 | TNM stage: OR =1.85, 95% CI: 0.80–4.25 | |||||||
| 6 | Depth of invasion: OR =2.49, 95% CI: 1.28–4.83 | |||||||
| 7 | Lymph node metastasis: OR =2.15, 95% CI: 1.27–3.66 | |||||||
| 3 | Distant metastasis: OR =3.26, 95% CI: 0.17–61.62 | |||||||
| 6 | Grade of differentiation: OR =1.87, 95% CI: 0.95–3.66 | |||||||
| 5 | Vascular invasion: OR =2.23, 95% CI: 1.20–4.14 |
Notes: *In the study by Li Y, based on the 95% CI of OR, the statistical difference should not be significant.
Table S11
| First author | Journal (Year) | No. studies | Included studies | No. Case | No. Control | Results | Model |
|---|---|---|---|---|---|---|---|
| Li Y | PLoS One [2013] | 1 | Li K, et al. Biochem Genet [2009] | 87 | 106 | CT vs. CC: OR =0.34, 95% CI: 0.11–1.10, P=0.072 | A fixed-effect model was used when P heterogeneity <0.05, otherwise a random effect model was used |
| TT/CT vs. CC: OR =0.34, 95% CI: 0.11–1.10, P=0.072 | |||||||
| T allele vs. C allele: OR =0.36, 95% CI: 0.12–1.13, P=0.079 | |||||||
| Xu J | Genet Test Mol Biomarkers [2013] | 1 | Li K, et al. Biochem Genet [2009] | 87 | 106 | C allele vs. T allele: OR =5.17, 95% CI: 1.75–15.26, P=0.003 | When a significant Q-test with P<0.05 or I2>50% indicated that heterogeneity among studies existed, the random effects model (DerSimonian Laird method) was conducted for the meta-analysis; otherwise, the fixed effects model (Mantel–Haenszel method) was used |
| CC vs. TT + CT: OR =5.75, 95% CI: 1.91–17.35, P=0.002 |
Table S12
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Anam MT | Biomark Res [2015] | Bangladesh | PubMed, PubMed Central, Google Scholar | 2014.12 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 6 | Risk: |
| TT vs. CC: OR =0.84, 95% CI: 0.47–1.49, P=0.5449 | ||||||||
| CT vs. CC: OR =1.34, 95% CI: 0.95–1.87, P=0.0913 | ||||||||
| TT + CT vs. CC: OR =1.33, 95% CI: 0.95–1.87, P=0.0982 | ||||||||
| TT vs. CT + CC: OR =0.81, 95% CI: 0.47–1.40, P=0.4535 | ||||||||
| T allele vs. C allele: OR =1.29, 95% CI: 0.94–1.76, P=0.1178 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| AA vs. GG: OR =3.35, 95% CI: 0.14–82.30, P=0.4597 | ||||||||
| GA vs. GG: OR =1.41, 95% CI: 0.96–2.08, P=0.0822 | ||||||||
| AA vs. GA + GG: OR =3.25, 95% CI: 0.13–79.90, P=0.4707 | ||||||||
| AA + GA vs. GG: OR =1.41, 95% CI: 0.93–2.15, P=0.1043 | ||||||||
| A allele vs. G allele: OR =1.42, 95% CI: 0.93–2.17, P=0.1093 | ||||||||
| Renal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: | |||||
| TT vs. CC: OR =0.27, 95% CI: 0.08–0.90, P=0.0335 | ||||||||
| CT vs. CC: OR =0.40, 95% CI: 0.12–1.34, P=0.1369 | ||||||||
| TT + CT vs. CC: OR =0.43, 95% CI: 0.15–1.20, P=0.1082 | ||||||||
| TT vs. CT + CC: OR =1.08, 95% CI: 0.44–2.64, P=0.8703 | ||||||||
| T allele vs. C allele: OR =0.84, 95% CI: 0.58–1.22, P=0.3548 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 4 | Risk: | ||||||
| AA vs. GG: OR =5.11, 95% CI: 2.24–11.66, P=0.0001 | ||||||||
| GA vs. GG: OR =1.51, 95% CI: 0.45–5.05, P=0.5038 | ||||||||
| AA vs. GA + GG: OR =3.05, 95% CI: 1.36–6.84, P=0.0068 | ||||||||
| AA + GA vs. GG: OR =1.58, 95% CI: 0.49–5.03, P=0.442 | ||||||||
| A allele vs. G allele: OR =1.53, 95% CI: 0.60–3.92, P=0.3747 | ||||||||
| Fan Y | Medicine [2015] | China | PubMed, Embase, Web of Science, Cochrane, EBSCO, CINAHL, Biological Abstracts | 2015.8.15 | Renal cell carcinoma | HIF-1α nuclear and cytoplasmic expression | 5 | OS: HR =1.637, 95% CI: 0.898–2.985, P=0.108 |
| 6 | Cancer-specific survival: HR=1.110, 95% CI: 0.595–2.069, P=0.744 | |||||||
| 4 | PFS: HR =1.113, 95% CI: 0.675–1.836, P=0.674 | |||||||
| HIF-2α nuclear and cytoplasmic expression | 4 | Cancer-specific survival: HR=1.597, 95% CI: 0.667–3.824, P=0.293 | ||||||
| 3 | PFS: HR =0.847, 95% CI: 0.566–1.266, P=0.417 | |||||||
| He P | PLoS One [2013] | China | PubMed, Embase, CNKI | 2013.8.23 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 6 | Dominant model (TT + CT vs. CC): OR =1.36, 95% CI: 0.95–1.96 | |||||||
| 5 | Recessive model (TT vs. CT + CC): OR =1.31, 95% CI: 0.54–3.18 | |||||||
| 5 | Homozygote comparison (TT vs. CC): OR =1.34, 95% CI: 0.54–3.30 | |||||||
| 6 | Heterozygote comparison (CT vs. CC): OR =1.34, 95% CI: 0.93–1.92 | |||||||
| Renal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | ||||||
| 3 | Dominant model (TT + CT vs. CC): OR = 0.46, 95% CI: 0.13–1.60 | |||||||
| 3 | Recessive model (TT vs. CT + CC): OR =1.55, 95% CI: 1.02–2.37 | |||||||
| 3 | Homozygote comparison (TT vs. CC): OR =0.29, 95% CI: 0.06–1.45 | |||||||
| 3 | Heterozygote comparison (CT vs. CC): OR =0.44, 95% CI: 0.11–1.69 | |||||||
| Hu X | Tumour Biol [2014] | China | PubMed, Embase, CNKI | 2013.7 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 5 | Risk: |
| T allele vs. C allele: OR =1.54, 95% CI: 1.04–2.30 | ||||||||
| TT vs. CC: OR =1.91, 95% CI: 0.82–4.47 | ||||||||
| CT vs. CC: OR =1.54, 95% CI: 0.95–2.49 | ||||||||
| TT + CT vs. CC: OR =1.58, 95% CI: 1.00–2.49 | ||||||||
| TT vs. CT + CC: OR =1.88, 95% CI: 0.79–4.47 | ||||||||
| Renal cell carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: | |||||
| T allele vs. C allele: OR =0.92, 95% CI: 0.70–1.19 | ||||||||
| TT vs. CC: OR =0.37, 95% CI: 0.12–1.12 | ||||||||
| CT vs. CC: OR =0.64, 95% CI: 0.32–1.29 | ||||||||
| TT + CT vs. CC: OR =0.65, 95% CI: 0.35–1.23 | ||||||||
| TT vs. CT + CC: OR =1.31, 95% CI: 0.77–2.24 | ||||||||
| Bladder cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: TT + CT vs. CC: OR =1.12, 95% CI: 0.65–1.92 | |||||
| Li D | PLoS One [2013] | China | PubMed | 2012.11.25 | Overall urinary cancers | HIF-1α gene P582S polymorphism | Risk: | |
| 11 | TT vs. CT + CC: OR = 1.17, 95% CI: 0.67–2.05, P=0.57 | |||||||
| 11 | TT + CT vs. CC: OR =1.10, 95% CI: 0.83–1.45, P=0.52 | |||||||
| 11 | T allele vs. C allele: OR =1.13, 95% CI: 0.90–1.41, P=0.30 | |||||||
| HIF-1α gene A588T polymorphism | Risk: | |||||||
| 9 | AA + AG vs. GG: OR =1.40, 95% CI: 0.76–2.58, P=0.28 | |||||||
| 8 | A allele vs. G allele: OR =1.57, 95% CI: 0.89–2.76, P=0.12 | |||||||
| Prostate cancer | HIF-1α gene P582S polymorphism | Risk: | ||||||
| 6 | TT vs. CT + CC: OR = 1.31, 95% CI: 0.54–3.20, P=0.55 | |||||||
| 6 | TT + CT vs. CC: OR =1.36, 95% CI: 0.95–1.96, P=0.09 | |||||||
| 6 | T allele vs. C allele: OR =1.35, 95% CI: 0.96–1.89, P=0.08 | |||||||
| HIF-1α gene A588T polymorphism | Risk: | |||||||
| 4 | AA + AG vs. GG: OR =1.45, 95% CI: 1.00–2.12, P=0.05 | |||||||
| 4 | A allele vs. G allele: OR =1.46, 95% CI: 1.01–2.12, P=0.04 | |||||||
| Renal cancer | HIF-1α gene P582S polymorphism | Risk: | ||||||
| 4 | TT vs. CT + CC: OR = 1.37, 95% CI: 0.92–2.04, P=0.12 | |||||||
| 4 | TT + CT vs. CC: OR =0.62, 95% CI: 0.33–1.19, P=0.15 | |||||||
| 4 | T allele vs. C allele: OR =0.91, 95% CI: 0.73–1.12, P=0.37 | |||||||
| HIF-1α gene A588T polymorphism | Risk: | |||||||
| 4 | AA + AG vs. GG: OR =1.58, 95% CI: 0.49–5.03, P=0.44 | |||||||
| 4 | A allele vs. G allele: OR =1.53, 95% CI: 0.60–3.92, P=0.38 | |||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| TT vs. CC: OR =2.02, 95% CI: 0.60–6.83, P=0.117 | ||||||||
| CT vs. CC: OR =1.42, 95% CI: 0.84–2.40, P=0.062 | ||||||||
| TT/CT vs. CC: OR =1.46, 95% CI: 0.89–2.40, P=0.031 | ||||||||
| TT vs. CT/CC: OR =2.03, 95% CI: 0.58–7.16, P=0.124 | ||||||||
| T allele vs. C allele: OR =1.43, 95% CI: 0.93–2.21, P=0.017 | ||||||||
| Renal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: | |||||
| TT vs. CC: OR =0.67, 95% CI: 0.21–2.15C0.498 | ||||||||
| CT vs. CC: OR =0.92, 95% CI: 0.67–1.26, P=0.599 | ||||||||
| TT/CT vs. CC: OR =0.90, 95% CI: 0.67–1.22, P=0.509 | ||||||||
| TT vs. CT/CC: OR =0.69, 95% CI: 0.22–2.17, P=0.521 | ||||||||
| T allele vs. C allele: OR =0.89, 95% CI: 0.67–1.19, P=0.432 | ||||||||
| Bladder cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 1 | Risk: | |||||
| CT vs. CC: OR =1.11, 95% CI: 0.65–1.92, P=0.697 | ||||||||
| TT/CT vs. CC: OR =1.11, 95% CI: 0.65–1.92, P=0.697 | ||||||||
| T allele vs. C allele: OR =1.11, 95% CI: 0.65–1.88, P=0.704 | ||||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Prostate cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: |
| AA vs. GG: OR =3.35, 95% CI: 0.14–82.30, P=0.460 | ||||||||
| GA vs. GG: OR =1.41, 95% CI: 0.97–2.07, P=0.082 | ||||||||
| AA + GA vs. GG: OR =1.44, 95% CI: 0.98–2.10, P=0.104 | ||||||||
| AA vs. GA + GG: OR =3.25, 95% CI: 0.13–79.90, P=0.471 | ||||||||
| A allele vs. G allele: OR =1.45, 95% CI: 1.00–2.11, P=0.109 | ||||||||
| Renal cell carcinoma | HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | |||||
| AA vs. GG: OR =4.70, 95% CI: 0.22–98.24, P=0.319 | ||||||||
| GA vs. GG: OR =1.00, 95% CI: 0.69–1.47, P=0.975 | ||||||||
| AA + GA vs. GG: OR =1.04, 95% CI: 0.71–1.51, P=0.841 | ||||||||
| AA vs. GA + GG: OR =4.78, 95% CI: 0.23–100.04, P=0.313 | ||||||||
| A allele vs. G allele: OR =1.07, 95% CI: 0.74–1.55, P=0.706 | ||||||||
| Tian Y | Chinese Journal of Evidence–Based Medicine [2015] Article in Chinese | China | Cochrane, PubMed, Embase, Ovid, CNKI, VIP, CBM, WanFang | 2015.6 | Renal cell cancer | HIF-1α expression | 7 | Risk: OR =16.76, 95% CI: 8.53–32.92, p<0.00001 |
| 7 | Lymph node metastasis: (yes vs. no): OR =4.33, 95% CI: 2.53–7.39, p<0.00001 | |||||||
| 5 | Clinical stage I–II vs. stage III–IV: OR =0.3, 95% CI: 0.18–0.51, p<0.0001 | |||||||
| 4 | Pathological stage G1+G2 vs. G3+G4: OR =0.54, 95% CI: 0.29–0.98, P=0.04 | |||||||
| 3 | Age (≥ 50 vs. <50): OR =1.09, 95% CI: 0.54–2.19, P=0.82 | |||||||
| 6 | Male vs. Female: OR =0.77, 95% CI: 0.48–1.25, P=0.29 | |||||||
| Wu G | Tumour Biol [2014] | China | PubMed, Embase, Google Scholar, Wanfang | 2013.6.10 | Renal carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 4 | TT + CT vs. CC: OR =0.62, 95% CI: 0.33–1.19, P=0.15 | |||||||
| 4 | TT vs. CT + CC: OR =0.96, 95% CI: 0.76–1.20, P=0.706 | |||||||
| Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | ||||||
| 6 | TT + CT vs. CC: OR =1.36, 95% CI: 0.95–1.96, P=0.094 | |||||||
| 6 | TT vs. CT + CC: OR =1.27, 95% CI: 0.93–1.73, P=0.126 | |||||||
| Yan Q | BMC Cancer [2014] | China | PubMed, Web of Science | 2013.9.20 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 6 | Risk: |
| TT vs. CC: OR =1.34, 95% CI: 0.54–3.31 | ||||||||
| CT vs. CC: OR =1.34, 95% CI: 0.93–1.92 | ||||||||
| TT + CT vs. CC: OR =1.36, 95% CI: 0.95–1.96 | ||||||||
| TT vs. CT + CC: OR =1.31, 95% CI: 0.54–3.20 | ||||||||
| T allele vs. C allele: OR =1.35, 95% CI: 0.96–1.89 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| GA vs. GG: OR =1.42, 95% CI: 0.97–2.07 | ||||||||
| AA + GA vs. GG: OR =1.44, 95% CI: 0.98–2.10 | ||||||||
| A allele vs. G allele: OR =1.45, 95% CI: 0.99–2.11 | ||||||||
| Renal cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: | |||||
| TT vs. CC: OR =0.28, 95% CI: 0.12–1.28 | ||||||||
| CT vs. CC: OR =0.62, 95% CI: 0.31–1.24 | ||||||||
| TT + CT vs. CC: OR =0.62, 95% CI: 0.33–1.18 | ||||||||
| TT vs. CT + CC: OR =1.37, 95% CI: 0.92–2.04 | ||||||||
| T allele vs. C allele: OR =0.91, 95% CI: 0.73–1.12 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 4 | Risk: | ||||||
| AA vs. GG: OR =5.10, 95% CI: 2.21–11.73 | ||||||||
| GA vs. GG: OR =1.51, 95% CI: 0.45–5.05 | ||||||||
| AA + GA vs. GG: OR =1.58, 95% CI: 0.49–5.04 | ||||||||
| AA vs. GA + GG: OR =3.09, 95% CI: 1.38–6.92 | ||||||||
| A allele vs. G allele: OR =1.53, 95% CI: 0.60–3.92 | ||||||||
| Yang X | PLoS One [2013] | China | PubMed, Embase | 2013.6.26 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 5 | Risk: |
| TT vs. CC: OR =3.68, 95% CI: 1.58–8.55 | ||||||||
| CT vs. CC: OR =2.02, 95% CI: 1.01–4.07 | ||||||||
| TT + CT vs. CC: OR =2.10, 95% CI: 1.08–4.09 | ||||||||
| TT vs. CT + CC: OR =3.52, 95% CI: 1.52–8.16 | ||||||||
| T allele vs. C allele: OR =2.06, 95% CI: 1.15–3.68 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| AA vs. GG: OR =3.35, 95% CI: 0.14–82.3 | ||||||||
| GA vs. GG: OR =1.41, 95% CI: 0.97–2.07 | ||||||||
| AA + GA vs. GG: OR =1.44, 95% CI: 0.98–2.10 | ||||||||
| AA vs. GA + GG: OR =3.25, 95% CI: 0.13–79.9 | ||||||||
| A allele vs. G allele: OR =1.45, 95% CI: 1.00–2.11 | ||||||||
| Ye Y | Cancer Invest [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 5 | Risk: TT + CT vs. CC: OR =1.59, 95% CI: 1.11–2.28, P=0.01 |
| Renal cell carcinoma | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: TT + CT vs. CC: OR =1.06, 95% CI: 0.41–2.73, P=0.9 | |||||
| Ye Y | Tumori [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Prostate cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 3* | Risk: TT + CT vs. CC: OR =0.98, 95% CI: 0.55–1.76, P=0.95* |
| Renal cell carcinoma | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: TT + CT vs. CC: OR =2.47, 95% CI: 0.21–28.92, P=0.47 | |||||
| Zhao T | J Exp Clin Cancer Res (2009) | China | PubMed | 2009.6 | Prostate cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| T allele vs. C allele: OR =1.78, 95% CI: 1.07–2.94, P=0.03 | ||||||||
| TT vs. CT + CC: OR =1.53, 95% CI: 0.90–2.60, P=0.11 | ||||||||
| TT + CT vs. CC: OR =1.85, 95% CI: 1.04–3.31, P=0.04 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: | ||||||
| A allele vs. G allele: OR =0.96, 95% CI: 0.49–1.90, P=0.91 | ||||||||
| AA + GA vs. GG: OR =0.96, 95% CI: 0.49–1.90, P=0.91 | ||||||||
| Zhou Y | Cancer Cell Int [2014] | China | PubMed, Embase, CNKI | 2013.12.13 | Prostate cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | |
| 3 | AA + GA vs. GG: OR =1.41, 95% CI: 0.93–2.14 | |||||||
| 1 | AA vs. GA + GG: OR =3.24, 95% CI: 0.13–79.9 | |||||||
| 1 | AA vs. GG: OR =3.34, 95% CI: 0.13–82.30 | |||||||
| 1 | GA vs. GG: OR =1.98, 95% CI: 0.07–50.4 | |||||||
| Renal cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | Risk: | ||||||
| 3 | AA + GA vs. GG: OR =0.94, 95% CI: 0.16–5.29 | |||||||
| 2 | AA vs. GA + GG: OR =2.69, 95% CI: 1.20–6.03 | |||||||
| 2 | AA vs. GG: OR =3.71, 95% CI: 1.72–7.99 | |||||||
| 1 | GA vs. GG: OR =0.81, 95% CI: 0.33–2 |
Notes: *In the study by Ye Y (Tumori, 2014), the number of included studies regarding prostate cancer should be 3, but not 4. Accordingly, the statistical results should not be reliable.
Table S13
| First author | Journal [year] | No. studies | Included studies | No. Case | No. Control | Results | Model |
|---|---|---|---|---|---|---|---|
| Anam MT | Biomark Res [2015] | 4 | Qin C, et al. Ann Oncol [2012] | 620 | 623 | AA vs. GG: OR =5.11, 95% CI: 2.24–11.66, P=0.0001; GA vs. GG: OR =1.51, 95% CI: 0.45–5.05, P=0.5038; AA vs. GA + GG: OR =3.05, 95% CI: 1.36–6.84, P=0.0068; AA + GA vs. GG: OR =1.58, 95% CI: 0.49–5.03, P=0.442; A allele vs. G allele: OR =1.53, 95% CI: 0.60–3.92, P=0.3747 |
Maybe a random-effects model was employed according to the forest plots |
| Morris MR, et al. Anticancer Res [2009] | 325 | 309 | |||||
| Ollerenshaw M, et al. Cancer Genet Cytogenet [2004] | 146 | 288 | |||||
| Clifford SC, et al. Oncogene [2001] | 48 | 144 | |||||
| Li D | PLoS One [2013] | 4 | Qin C, et al. Ann Oncol [2012] | 620 | 623 | AA + AG vs. GG: OR =1.58, 95% CI: 0.49–5.03, P=0.44; A allele vs. G allele: OR =1.53, 95% CI: 0.60–3.92, P=0.38 |
The random-effects model (the Dersimonian-Laird method) would be used if the test of heterogeneity was significant; otherwise the fixed-effects model (the Mantel-Haenszel method) would be applied in the analysis |
| Morris MR, et al. Anticancer Res [2009] | 325 | 309 | |||||
| Ollerenshaw M, et al. Cancer Genet Cytogenet [2004] | 146 | 288 | |||||
| Clifford SC, et al. Oncogene [2001] | 48 | 144 | |||||
| Yan Q | BMC Cancer [2014] | 4 | Qin C, et al. Ann Oncol [2012] | 620 | 623 | AA vs. GG: OR =5.10, 95% CI: 2.21–11.73; GA vs. GG: OR =1.51, 95% CI: 0.45–5.05; AA vs. GA + GG: OR =3.09, 95% CI: 1.38–6.92; AA + GA vs. GG: OR =1.58, 95% CI: 0.49–5.04; A allele vs. G allele: OR =1.53, 95% CI: 0.60–3.92 |
When P>0.05, the effects were assumed to be homogenous, and the fixed-effect model (the Mantel-Haenszel method) was used. When P<0.05, the random-effect model (the DerSimonian and Laird method) was more appropriate |
| Morris MR, et al. Anticancer Res [2009] | 325 | 309 | |||||
| Ollerenshaw M, et al. Cancer Genet Cytogenet [2004] | 146 | 288 | |||||
| Clifford SC, et al. Oncogene [2001] | 48 | 144 |
Table S14
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| He P | PLoS One [2013] | China | PubMed, Embase, CNKI | 2013.8.23 | Cervical cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | Risk: | |
| 3 | Dominant model (TT + CT vs. CC): OR =1.81, 95% CI: 0.79–4.10 | |||||||
| 2 | Recessive model (TT vs. CT + CC): OR =8.80, 95% CI: 2.31–33.52 | |||||||
| 2 | Homozygote comparison (TT vs. CC): OR =11.49, 95% CI: 2.21–59.67 | |||||||
| 3 | Heterozygote comparison (CT vs. CC): OR =1.47, 95% CI: 0.79–2.74 | |||||||
| Hu X | Tumour Biol [2013] | China | PubMed, Embase, CNKI | 2013.2 | Cervical cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Lymph node metastasis: OR =1.32, 95% CI: 0.60–2.90, P=0.493 |
| Hu X | Tumour Biol [2014] | China | PubMed, Embase, CNKI | 2013.7 | Cervical cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| T allele vs. C allele: OR =1.89, 95% CI: 0.84–4.26 | ||||||||
| TT vs. CC: OR =11.49, 95% CI: 2.18–60.52 | ||||||||
| CT vs. CC: OR =1.47, 95% CI: 0.79–2.74 | ||||||||
| TT + CT vs. CC: OR =1.81, 95% CI: 0.79–4.11 | ||||||||
| TT vs. CT + CC: OR =8.80, 95% CI: 2.30–33.70 | ||||||||
| Huang M | Int J Gynecol Cancer [2014] | China | Medline, PubMed, Embase, Web of Science | 2013.1 | Cervical cancer | HIF-1α expression | 7 | DFS: HR =1.98, 95% CI: 1.22–3.21, P=0.006 |
| 7 | OS: HR =2.58, 95% CI: 1.86–3.56, P<0.001 | |||||||
| 5 | Lymph node metastasis (yes vs no): OR =2.58, 95% CI: 1.86–3.56, P=0.167 | |||||||
| 3 | Tumor grade (grade 3 vs. grade 1/2): OR =0.99, 95% CI: 0.54–1.82, P=0.969 | |||||||
| 5 | Tumor size (size≥4 cm vs. size <4 cm): OR =2.04, 95% CI: 1.24–3.34, P=0.005 | |||||||
| 4 | FIGO stage (advanced stage vs. earlier stage): OR =1.52, 95% CI: 0.87–2.69, P=0.145 | |||||||
| 4 | Histology type (other type vs. SCC): OR =1.63, 95% CI: 0.85–3.13, P=0.139 | |||||||
| 3 | Anemia (yes vs. no): OR =2.04, 95% CI: 1.07–3.88, P=0.030 | |||||||
| Jin Y | Tumour Biol [2014] | China | PubMed, Cochrane, Web of Science, CNKI | 2014.2 | Epithelial ovarian cancer | HIF-1α expression | 3 | 5-year survival rate: OR =11.46, 95% CI: 3.43–38.29, P<0.0001 |
| Pathological type: | ||||||||
| 13 | Cancer vs. benign: OR =9.73, 95% CI: 4.90–19.32, P<0.00001 | |||||||
| 10 | Cancer vs. borderline: OR =2.31, 95% CI: 1.04–5.09, P=0.04 | |||||||
| 9 | Borderline vs. benign: OR =6.29, 95% CI: 2.69–14.73, P<0.0001 | |||||||
| NA | Histological type: | |||||||
| Serous vs. others: OR =1.02, 95% CI: 0.79–1.31, P=0.88 | ||||||||
| Serous vs. others: OR =1.37, 95% CI: 0.78–2.42, P=0.28 | ||||||||
| 17 | FIGO (III–IV vs. I–II): OR =3.01, 95% CI: 1.92–4.74, P<0.00001 | |||||||
| 10 | Histological grade: | |||||||
| Grades G3 vs. G1: OR =4.52, 95% CI: 2.79–7.31, P<0.00001 | ||||||||
| Grades G3 vs. G2: OR =2.02, 95% CI: 1.27–3.19, P=0.003 | ||||||||
| Grades G2 vs. G1: OR =2.43, 95% CI: 1.65–3.59, P<0.0001 | ||||||||
| 9 | Lymph node metastasis: OR =5.20, 95% CI: 2.10–12.89, P=0.0004 | |||||||
| Jin Y | PLoS One [2015] | China | PubMed, Cochrane, Web of Knowledge, clinical trial registries | 2014.1 | Overall gynecological cancer | HIF-1α expression | 9 | 5–year DFS rate: OR =2.93, 95% CI: 1.43–6.01, P=0.003 |
| 8 | 5–year OS rate: OR =5.53, 95% CI: 2.48–12.31, P<0.0001 | |||||||
| Pathological type: | ||||||||
| 21 | Cancer vs. Borderline: OR =2.70, 95% CI: 1.69–4.31, P<0.0001 | |||||||
| 26 | Cancer vs. Normal: OR =9.59, 95% CI: 5.97–15.39, P<0.00001 | |||||||
| 19 | Borderline vs. Normal: OR =4.13, 95% CI: 2.43–7.02, P<0.00001 | |||||||
| 32 | FIGO stage: OR =2.66, 95% CI: 1.87–3.79, P<0.00001 | |||||||
| Histological type: | ||||||||
| 22 | G3 vs. G1: OR =3.77, 95% CI: 2.76–5.16, P<0.00001 | |||||||
| 22 | G3 vs. G2: OR =1.62, 95% CI: 1.20–2.19, P=0.002 | |||||||
| 22 | G2 vs. G1:OR =2.34, 95% CI: 1.82–3.00, P<0.00001 | |||||||
| 21 | Lymph node metastasis: OR =3.98, 95% CI: 2.10–12.89, P<0.0001 | |||||||
| Endometrial cancer | HIF-1α expression | 4 | 5-year DFS rate: OR =1.56, 95% CI: 0.36–6.83, P=0.55 | |||||
| 2 | 5-year OS rate: OR =3.67, 95% CI: 0.52–25.63, P=0.19 | |||||||
| Pathological type: | ||||||||
| 4 | Cancer vs. Borderline: OR =4.45, 95% CI: 2.57–7.71, P<0.00001 | |||||||
| 6 | Cancer vs. Normal: OR =11.03, 95% CI: 6.55–18.58, P<0.00001 | |||||||
| 3 | Borderline vs. Normal: OR =3.48, 95% CI: 0.75–16.15, P=0.11 | |||||||
| 11 | FIGO stage: OR =2.76, 95% CI: 1.25–6.09, P=0.01 | |||||||
| Histological type: | ||||||||
| 6 | G3 vs. G1: OR =2.65, 95% CI: 1.53–4.59, P=0.0005 | |||||||
| 6 | G3 vs. G2: OR =1.15, 95% CI: 0.65–2.01, P=0.63 | |||||||
| 6 | G2 vs. G1: OR =2.19, 95% CI: 1.43–3.37, P=0.0003 | |||||||
| 4 | Lymph node metastasis: OR =4.02, 95% CI: 1.32–12.26, P=0.01 | |||||||
| Cervical cancer | HIF-1α expression | 3 | 5-year DFS rate: OR =5.28, 95% CI: 2.90–9.63, P<0.00001 | |||||
| 3 | 5-year OS rate: OR =3.28, 95% CI: 1.63–6.60, P=0.008 | |||||||
| Pathological type: | ||||||||
| 7 | Cancer vs. borderline: OR =2.36, 95% CI: 1.04–5.38, P=0.04 | |||||||
| 7 | Cancer vs. normal: OR =8.17, 95% CI: 2.80–23.85, P=0.0001 | |||||||
| 7 | Borderline vs. normal: OR =2.40, 95% CI: 1.52–3.78, P=0.0002 | |||||||
| 4 | FIGO stage: | |||||||
| OR =1.76, 95% CI: 1.03–2.99, P=0.04 (fixed-effect model) | ||||||||
| OR =1.69, 95% CI: 0.90–3.15, P=0.10 (random-effect model) | ||||||||
| Histological type: | ||||||||
| 6 | G3 vs. G1: OR =4.29, 95% CI: 2.26–8.14, P<0.00001 | |||||||
| 6 | G3 vs. G2: OR =1.62, 95% CI: 0.91–2.90, P=0.10 | |||||||
| 6 | G2 vs. G1: OR =2.40, 95% CI: 1.46–3.93, P=0.0005 | |||||||
| 8 | Lymph node metastasis: OR =2.94, 95% CI: 1.19–7329, P=0.02 | |||||||
| Ovarian cancer | HIF-1α expression | 2 | 5-year DFS rate: OR =2.42, 95% CI: 0.80–7.36, P=0.12 | |||||
| 3 | 5-year OS rate: OR =11.46, 95% CI: 3.43–38.29, P<0.0001 | |||||||
| Pathological type: | ||||||||
| 10 | Cancer vs. borderline: OR =2.31, 95% CI: 1.04–5.09, P=0.04 | |||||||
| 13 | Cancer vs. normal: OR =9.73, 95% CI: 4.90–19.32, P<0.00001 | |||||||
| 9 | Borderline vs. normal: OR =6.29, 95% CI: 2.69–14.73, P<0.0001 | |||||||
| 17 | FIGO stage: OR =3.01, 95% CI: 1.92–4.74, P<0.00001 | |||||||
| Histological type: | ||||||||
| 10 | G3 vs. G1: OR =4.52, 95% CI: 2.79–7.31, P<0.00001 | |||||||
| 10 | G3 vs. G2: OR =2.02, 95% CI: 1.27–3.19, P=0.003 | |||||||
| 10 | G2 vs. G1: OR =2.43, 95% CI: 1.65–3.59, P<0.00001 | |||||||
| 9 | Lymph node metastasis: OR =5.20, 95% CI: 2.10–12.89, P=0.0004 | |||||||
| Li Y | Int J Clin Exp Med [2015] | China | PubMed, Web of Knowledge, Medline, Embase, Google Scholar | 2014.7 | Gynecological cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 2 | Risk: |
| TT vs. CC: OR =9.92, 95% CI: 2.15–45.66, P=0.003 | ||||||||
| CT vs. CC: OR =1.16, 95% CI: 0.77–1.75, P=0.488 | ||||||||
| TT/CT vs. CC: OR =1.31, 95% CI: 0.58–2.94, P=0.152 | ||||||||
| TT vs. CT/CC: OR =8.35, 95% CI: 1.85–37.75, P=0.006 | ||||||||
| T allele vs. C allele: OR =1.38, 95% CI: 0.58–3.29, P=0.020 | ||||||||
| Liu P | Neoplasma [2014] | China | PubMed, Embase, Web of Knowledge, Google Scholar | 2013.8 | Gynecological cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: |
| AA vs. GG: OR =0.36, 95% CI: 0.01–8.80, P=0.528 | ||||||||
| GA vs. GG: OR =1.16, 95% CI: 0.54–2.48, P=0.744 | ||||||||
| AA + GA vs. GG: OR =1.08, 95% CI: 0.51–2.28, P=0.791 | ||||||||
| AA vs. GA + GG: OR =0.36, 95% CI: 0.01–8.81, P=0.529 | ||||||||
| A allele vs. G allele: OR =1.00, 95% CI: 0.48–2.08, P=0.831 | ||||||||
| Sun C | Ai Zheng. Ji Bian. Tu Bian. [2015]; Article in Chinese | China | CNKI, CBM | 2014.3.10 | Epithelial ovarian cancer | HIF-1α protein expression | 6 | Risk: OR =0.036, 95% CI: 0.010–0.135, P<0.001 |
| 3 | Lymph node: OR =0.080, 95% CI: 0.029–0.220, P<0.001 | |||||||
| 6 | Clinical stage: OR =0.258, 95% CI: 0.136–0.490, P<0.001 | |||||||
| 4 | Pathological type: OR =1.779, 95% CI: 0.876–3.616, P=0.111 | |||||||
| 6 | Pathological stage: OR =0.327, 95% CI: 0.084–1.268, P=0.106 | |||||||
| 3 | Age: OR =1.331, 95% CI: 0.341–5.196, P=0.681 | |||||||
| Yan Q | BMC Cancer [2014] | China | PubMed, Web of Science | 2013.9.20 | Cervical cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| TT vs. CC: OR =10.11, 95% CI: 2.55–40.05 | ||||||||
| CT vs. CC: OR =0.98, 95% CI: 0.72–1.34 | ||||||||
| TT + CT vs. CC: OR =1.32, 95% CI: 0.61–2.87 | ||||||||
| TT vs. CT + CC: OR =8.55, 95% CI: 2.28–32.13 | ||||||||
| T allele vs. C allele: OR =1.41, 95% CI: 0.59–3.35 | ||||||||
| HIF-1α rs11549467 (1790 G/A) polymorphism | 3 | Risk: | ||||||
| AA vs. GG: OR =0.35, 95% CI: 0.04–3.39 | ||||||||
| GA vs. GG: OR =0.62, 95% CI: 0.40–0.98 | ||||||||
| AA + GA vs. GG: OR =0.60, 95% CI: 0.38–0.94 | ||||||||
| AA vs. GA + GG: OR =0.36, 95% CI: 0.04–3.450 | ||||||||
| A allele vs. G allele: OR =0.59, 95% CI: 0.38–0.91 | ||||||||
| Yang X | PLoS One [2013] | China | PubMed, Embase | 2013.6.26 | Cervical cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: |
| TT vs. CC: OR =10.1, 95% CI: 3.12–32.6 | ||||||||
| CT vs. CC: OR =1.37, 95% CI: 0.92–2.02 | ||||||||
| TT + CT vs. CC: OR =1.63, 95% CI: 1.12–2.37 | ||||||||
| TT vs. CT + CC: OR =8.26, 95% CI: 2.64–25.9 | ||||||||
| T allele vs. C allele: OR =1.89, 95% CI: 0.84–4.26 | ||||||||
| Ye Y | Cancer Invest [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Cervical cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 3 | Risk: TT + CT vs. CC: OR =1.78, 95% CI: 0.76, 4.18, P=0.18 |
| Ye Y | Tumori [2014] | China | Medline, Embase, Web of Science | 2012.2.20 | Cervical cancer | HIF-1α rs11549467 (1790 G/A) polymorphism | 2 | Risk: TT + CT vs. CC: OR =0.92, 95% CI: 0.41–2.03, P=0.83 |
| Zhu J | Int J Clin Exp Pathol [2014] | China | PubMed, Embase | 2014.1.10 | Cervical cancer | HIF-1α rs11549465 (1772 C/T) polymorphism | 4 | Risk: |
| TT vs. CC: OR =6.32, 95% CI: 2.28–17.55 | ||||||||
| CT vs. CC: OR =1.05, 95% CI: 0.80–1.38 | ||||||||
| TT + CT vs. CC: OR =1.13, 95% CI: 0.87–1.47 | ||||||||
| TT vs. CT + CC: OR =5.86, 95% CI: 2.13–16.11 |
Table S15
| First author | Journal [year] | Country | Databases | Search date | Cancer | HIF | No. studies | Results |
|---|---|---|---|---|---|---|---|---|
| Ren HY | Onco Targets Ther [2016] | China | PubMed, Embase, Web of Science | 2015.8.1 | Osteosarcoma | HIF-1α expression | 2 | OS: HR=3.0, 95% CI: 1.46–6.15, P=0.003 |
| 3 | DFS: HR=2.23, 95% CI: 1.26–3.92, P=0.006 | |||||||
| 5 | Metastasis (yes vs. no): OR =5.06, 95% CI: 2.87–8.92, P<0.00001 | |||||||
| 2 | Pathologic grade (high vs. low): OR =21.33, 95% CI: 4.60-98.88, P<0.0001 | |||||||
| 4 | Tumor stage (high vs. low): OR =10.29, 95% CI: 3.55-29.82, P<0.0001 | |||||||
| 2 | Chemotherapy response (poor vs. good): OR =9.68, 95% CI: 1.87–50.18, P=0.007 | |||||||
| 4 | Tumor size (large vs. small): OR =1.12, 95% CI: 0.22–5.76, P=0.89 | |||||||
| 3 | Tumor site (tibia or femur vs. elsewhere): OR =2.02, 95% CI: 0.10–39.71, P=0.46 | |||||||
| 2 | Histopathology (osteoblastic vs. other types): OR =0.70, 95% CI: 0.28–1.73, P=0.46 |
Acknowledgements
Funding: This study was partially supported by the grants from the National Natural Science Foundation of China (no. 81500474) for Dr. X Qi.
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2017.04.08). Xingshun Qi serves as an Editor-in-Chief of AME Medical Journal. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dhani N, Fyles A, Hedley D, et al. The clinical significance of hypoxia in human cancers. Semin Nucl Med 2015;45:110-21. [Crossref] [PubMed]
- Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev 2007;26:225-39. [Crossref] [PubMed]
- Jubb AM, Buffa FM, Harris AL. Assessment of tumour hypoxia for prediction of response to therapy and cancer prognosis. J Cell Mol Med 2010;14:18-29. [Crossref] [PubMed]
- Vaupel P. The role of hypoxia-induced factors in tumor progression. Oncologist 2004;9:10-7. [Crossref] [PubMed]
- Semenza GL, Nejfelt MK, Chi SM, et al. Hypoxia-inducible nuclear factors bind to an enhancer element located 3' to the human erythropoietin gene. Proc Natl Acad Sci U S A 1991;88:5680-4. [Crossref] [PubMed]
- Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev 1997;11:72-82. [Crossref] [PubMed]
- Ema M, Taya S, Yokotani N, et al. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U S A 1997;94:4273-8. [Crossref] [PubMed]
- Gu YZ, Moran SM, Hogenesch JB, et al. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr 1998;7:205-13. [PubMed]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003;3:721-32. [Crossref] [PubMed]
- Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1). Mol Pharmacol 2006;70:1469-80. [Crossref] [PubMed]
- Anam MT, Ishika A, Hossain MB, et al. A meta-analysis of hypoxia inducible factor 1-alpha (HIF1A) gene polymorphisms: association with cancers. Biomark Res 2015;3:29. [Crossref] [PubMed]
- He P, Han Q, Liu J, et al. The association between hypoxia-inducible factor-1 alpha gene C1772T polymorphism and cancer risk: a meta-analysis of 37 case-control studies. PLoS One 2013;8:e83441. [Crossref] [PubMed]
- Hu X, Lin S, Zheng J, et al. Clinicopathological significance of hypoxia-inducible factor-1 alpha polymorphisms in cancers: evidence from a meta-analysis. Tumour Biol 2013;34:2477-87. [Crossref] [PubMed]
- Li Y, Li C, Shi H, et al. The association between the rs11549465 polymorphism in the hif-1alpha gene and cancer risk: a meta-analysis. Int J Clin Exp Med 2015;8:1561-74. [PubMed]
- Liu J, Zhang HX. 1790 G/A polymorphism, but not 1772 C/T polymorphism, is significantly associated with cancers: an update study. Gene 2013;523:58-63. [Crossref] [PubMed]
- Liu P, Shi H, Yang Y, et al. Update meta-analysis on 1790G/A polymorphism and cancer risk: Evidence from 26 studies. Neoplasma 2014;61:340-51. [Crossref] [PubMed]
- Wu G, Yan WF, Zhu YZ, et al. Hypoxia-inducible factor-1alpha (HIF-1alpha) C1772T polymorphism significantly contributes to the risk of malignancy from a meta-analysis. Tumour Biol 2014;35:4113-22. [Crossref] [PubMed]
- Yang X, Zhu HC, Zhang C, et al. HIF-1alpha 1772 C/T and 1790 G/A polymorphisms are significantly associated with higher cancer risk: an updated meta-analysis from 34 case-control studies. PLoS One 2013;8:e80396. [Crossref] [PubMed]
- Ye Y, Wang M, Hu S, et al. Hypoxia-inducible factor-1alpha C1772T polymorphism and cancer risk: a meta-analysis including 18,334 subjects. Cancer Invest 2014;32:126-35. [Crossref] [PubMed]
- Ye Y, Wang M, Li J, et al. Hypoxia-inducible factor-1alpha G polymorphism and the risk of cancer: a meta-analysis. Tumori 2014;100:e257-65. [PubMed]
- Zhang Q, Chen Y, Zhang B, et al. Hypoxia-inducible factor-1alpha polymorphisms and risk of cancer metastasis: a meta-analysis. PLoS One 2013;8:e70961. [Crossref] [PubMed]
- Zhao T, Lv J, Zhao J, et al. Hypoxia-inducible factor-1alpha gene polymorphisms and cancer risk: a meta-analysis. J Exp Clin Cancer Res 2009;28:159. [Crossref] [PubMed]
- Zhou Y, Lin L, Wang Y, et al. The association between hypoxia-inducible factor-1 alpha gene G1790A polymorphism and cancer risk: a meta-analysis of 28 case-control studies. Cancer Cell Int 2014;14:37. [Crossref] [PubMed]
- Liu Q, Cao P. Clinical and prognostic significance of HIF-1alpha in glioma patients: a meta-analysis. Int J Clin Exp Med 2015;8:22073-83. [PubMed]
- Hu X, Fang Y, Zheng J, et al. The association between HIF-1alpha polymorphism and cancer risk: a systematic review and meta-analysis. Tumour Biol 2014;35:903-16. [Crossref] [PubMed]
- Qian J, Wenguang X, Zhiyong W, et al. Hypoxia inducible factor: a potential prognostic biomarker in oral squamous cell carcinoma. Tumour Biol 2016;37:10815-20. [Crossref] [PubMed]
- Sun X, Liu YD, Gao W, et al. HIF-1alpha -1790G>A polymorphism significantly increases the risk of digestive tract cancer: a meta-analysis. World J Gastroenterol 2015;21:1641-9. [Crossref] [PubMed]
- Yan Q, Chen P, Wang S, et al. Association between HIF-1alpha C1772T/G1790A polymorphisms and cancer susceptibility: an updated systematic review and meta-analysis based on 40 case-control studies. BMC Cancer 2014;14:950. [Crossref] [PubMed]
- Yang X, Zhang C, Zhu HC, et al. HIF-1alpha P582S and A588T polymorphisms and digestive system cancer risk-a meta-analysis. Tumour Biol 2014;35:2825-30. [Crossref] [PubMed]
- Rainsbury JW, Ahmed W, Williams HK, et al. Prognostic biomarkers of survival in oropharyngeal squamous cell carcinoma: Systematic review and meta-analysis. Head and Neck 2013;35:1048-55. [Crossref] [PubMed]
- Jing SW, Zhao ZJ, Jing SH, et al. Relationship between hypoxia inducible factor-1α and clinicopathological features of nasopharyngeal carcinoma: A Meta analysis. Chinese Journal of Cancer Prevention and Treatment 2015;22:548-54.
- Li C, Lu HJ, Na FF, et al. Prognostic role of hypoxic inducible factor expression in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev 2013;14:3607-12. [Crossref] [PubMed]
- Liao SH, Liu WZ, Liu T, et al. Potential signaling pathway of hypoxia-inducible factor in lung cancer and its gene polymorphism with lung cancer risk. J Recept Signal Transduct Res 2015;35:233-7. [Crossref] [PubMed]
- Ren W, Mi D, Yang K, et al. The expression of hypoxia-inducible factor-1alpha and its clinical significance in lung cancer: a systematic review and meta-analysis. Swiss Med Wkly 2013;143:w13855. [PubMed]
- Wang Q, Hu DF, Rui Y, et al. Prognosis value of HIF-1alpha expression in patients with non-small cell lung cancer. Gene 2014;541:69-74. [Crossref] [PubMed]
- Ren HT, Wang XJ, Kang HF, et al. Associations between C1772T polymorphism in hypoxia-inducible factor-1alpha gene and breast cancer: a meta-analysis. Med Sci Monit 2014;20:2578-83. [Crossref] [PubMed]
- Sun G, Wang Y, Hu W. Correlation between HIF-1alpha expression and breast cancer risk: a meta-analysis. Breast J 2014;20:213-5. [Crossref] [PubMed]
- Wang W, He YF, Sun QK, et al. Hypoxia-inducible factor 1alpha in breast cancer prognosis. Clin Chim Acta 2014;428:32-7. [Crossref] [PubMed]
- Yin W, Liu G, Lu J, et al. Association of hypoxia-inducible factor-1 with breast cancer risk: A meta-analysis of published studies. Cancer Research 2011;71.
- Ni ZH, Liang XJ, Mo JG, et al. Associations of hypoxia inducible factor-1α gene polymorphisms with susceptibility to digestive tract cancers: a case–control study and meta-analysis. Genes and Genomics 2015;37:931-8. [Crossref]
- Xu JJ, Zou LY, Yang L, et al. Common polymorphisms in the HIF-1alpha gene confer susceptibility to digestive cancer: a meta-analysis. Genet Mol Res 2014;13:6228-38. [Crossref] [PubMed]
- Xu J, Xu L, Li L, et al. HIF-1alpha C1772T polymorphism and gastrointestinal tract cancer risk: a meta-analysis and meta-regression analysis. Genet Test Mol Biomarkers 2013;17:918-25. [Crossref] [PubMed]
- Xu J, Xu L, Li LT, et al. HIF1A gene Pro582Ser polymorphism and susceptibility to digestive tract cancers: a meta-analysis of case-control studies. Genet Mol Res 2014;13:5732-44. [Crossref] [PubMed]
- Chen Z, He X, Xia W, et al. Prognostic value and clinicopathological differences of HIFs in colorectal cancer: evidence from meta-analysis. PLoS One 2013;8:e80337. [Crossref] [PubMed]
- Cao S, Yang S, Wu C, et al. Protein expression of hypoxia-inducible factor-1 alpha and hepatocellular carcinoma: a systematic review with meta-analysis. Clin Res Hepatol Gastroenterol 2014;38:598-603. [Crossref] [PubMed]
- Chen J, Li T, Liu Q, et al. Clinical and prognostic significance of HIF-1alpha, PTEN, CD44v6, and survivin for gastric cancer: a meta-analysis. PLoS One 2014;9:e91842. [Crossref] [PubMed]
- Jing S, Wang J, Liu Q, et al. Relationship between hypoxia inducible factor-1alpha and esophageal squamous cell carcinoma: a meta analysis. Zhonghua Bing Li Xue Za Zhi 2014;43:593-9. [PubMed]
- Lin S, Ma R, Zheng XY, et al. Meta-analysis of immunohistochemical expression of hypoxia inducible factor-1alpha as a prognostic role in gastric cancer. World J Gastroenterol 2014;20:1107-13. [Crossref] [PubMed]
- Ping W, Sun W, Zu Y, et al. Clinicopathological and prognostic significance of hypoxia-inducible factor-1alpha in esophageal squamous cell carcinoma: a meta-analysis. Tumour Biol 2014;35:4401-9. [Crossref] [PubMed]
- Sun G, Hu W, Zhang J, et al. A meta-analysis of relationship between HIF-1α expression and esophageal squamous cell carinoma. J Chin Oncol 2012;18:686-91.
- Ye LY, Zhang Q, Bai XL, et al. Hypoxia-inducible factor 1alpha expression and its clinical significance in pancreatic cancer: a meta-analysis. Pancreatology 2014;14:391-7. [Crossref] [PubMed]
- Zhang ZG, Zhang QN, Wang XH, et al. Hypoxia-inducible factor 1 alpha (HIF-1alpha) as a prognostic indicator in patients with gastric tumors: a meta-analysis. Asian Pac J Cancer Prev 2013;14:4195-8. [Crossref] [PubMed]
- Zheng SS, Chen XH, Yin X, et al. Prognostic significance of HIF-1alpha expression in hepatocellular carcinoma: a meta-analysis. PLoS One 2013;8:e65753. [Crossref] [PubMed]
- Zhu CL, Huang Q, Liu CH, et al. Prognostic value of HIF-1alpha expression in patients with gastric cancer. Mol Biol Rep 2013;40:6055-62. [Crossref] [PubMed]
- Yao Q, Lv Y, Pan T, et al. Prognostic significance and clinicopathological features of hypoxic inducible factor-2alpha expression in hepatocellular carcinoma. Saudi Med J 2015;36:170-5. [Crossref] [PubMed]
- Zheng F, Du F, Zhao J. Clinicopathological Differences and Prognostic Value of Hypoxia-Inducible Factor-2alpha Expression for Gastric Cancer: Evidence From Meta-Analysis. Medicine (Baltimore) 2016;95:e2871. [Crossref] [PubMed]
- Li K, Zhang Y, Dan Z, et al. Association of the hypoxia inducible factor-1alpha gene polymorphisms with gastric cancer in Tibetans. Biochem Genet 2009;47:625-34. [Crossref] [PubMed]
- Fan Y, Li H, Ma X, et al. Prognostic Significance of Hypoxia-Inducible Factor Expression in Renal Cell Carcinoma: A PRISMA-compliant Systematic Review and Meta-Analysis. Medicine (Baltimore) 2015;94:e1646. [Crossref] [PubMed]
- Li D, Liu J, Zhang W, et al. Association between HIF1A P582S and A588T polymorphisms and the risk of urinary cancers: a meta-analysis. PLoS One 2013;8:e63445. [Crossref] [PubMed]
- Tian YJ, Guo Q, Chen ZH, et al. Correlation between HIF-1α protein expression and renal cell cancer risk: A meta-analysis. Chinese Journal of Evidence-Based Medicine 2015;15:1035-41.
- Huang M, Chen Q, Xiao J, et al. Overexpression of hypoxia-inducible factor-1alpha is a predictor of poor prognosis in cervical cancer: a clinicopathologic study and a meta-analysis. Int J Gynecol Cancer 2014;24:1054-64. [Crossref] [PubMed]
- Jin Y, Wang H, Liang X, et al. Pathological and prognostic significance of hypoxia-inducible factor 1alpha expression in epithelial ovarian cancer: a meta-analysis. Tumour Biol 2014;35:8149-59. [Crossref] [PubMed]
- Sun C, Jing S, Wang J, et al. Relationship between hypoxia inducible factor-1α and clinicopathology of epithelial ovarian cancer: Meta-analysis. Carcinogenesis, Teratogenesis & Mutagenesis 2015;27:49-53, 58.
- Zhu J, Cheng X, Xie R, et al. Genetic association between the HIF-1alpha P582S polymorphism and cervical cancer risk: a meta analysis. Int J Clin Exp Pathol 2014;7:6085-90. [PubMed]
- Jin Y, Wang H, Ma X, et al. Clinicopathological characteristics of gynecological cancer associated with hypoxia-inducible factor 1alpha expression: a meta-analysis including 6,612 subjects. PLoS One 2015;10:e0127229. [Crossref] [PubMed]
- Ren HY, Zhang YH, Li HY, et al. Prognostic role of hypoxia-inducible factor-1 alpha expression in osteosarcoma: a meta-analysis. Onco Targets Ther 2016;9:1477-87. [Crossref] [PubMed]
Cite this article as: Zou D, Han T, Deng H, Shao X, Guo X, Qi X. Hypoxia-inducible factors in cancer: an overview of major findings from meta-analyses. AME Med J 2017;2:48.