The association between let-7 microRNA-binding site polymorphism rs712 and cancer risk: a meta-analysis and trial sequential analysis
Introduction
MicroRNAs (miRNAs) are short sequences of small non-protein coding mRNA, which could bind to specific regions of target mRNA transcripts. MiRNAs post-transcriptionally regulate the expression level of the target gene in the way of inducing mRNA degradation or inhibiting mRNA translation, which depends on the matching degree between the miRNA and its target mRNA (1). Disregulation of miRNAs may function as both tumor suppressors and oncogenes (2,3). As a polymorphism at or near a miRNA binding site of functional genes, MiRNA-related polymorphism could influence gene expression by interfering with miRNA function (4). Recent studies have shown that single nucleotide polymorphisms (SNPs) residing on miRNA and/or miRNA-binding sites might be associated with human cancers (5,6).
As a member of ras gene family, The KRAS gene plays important roles in nosogenesis of cancer (7). Studies found the let-7 family of miRNAs binding to the 3′ untranslated region (UTR) of human KRAS could affect the binding ability and impact the following KRAS transcription (8). Previous studies have reported that let-7 binding site polymorphism rs712 increased cancer risk involving gastric cancer (GC), colorectal cancer (CRC), non-small cell carcinoma (NSCLC), oral squamous cell carcinoma (OSCC) and others, but some other studies observed insignificant or opposite results (5,9-13).
In order to drive a more precise relationship between rs712 and the risk of cancer, we thereby performed such meta-analysis.
Methods
Studies were searched in the electronic databases EMBASE, PubMed and Web of Science up to September 1st, 2016. Available publications were identified using the following keywords or text words: “rs712”, “let-7”, or “KRAS”, and “single nucleotide polymorphism”, or “polymorphism” and “cancer” or “carcinoma”. All studies were assessed to retrieve the most eligible literatures and the search was restricted on human subjects only. Besides, we also performed a manual search for relevant researches from reference lists by internet search engines. We also inspected review articles to search other eligible studies.
The included studies must met the inclusion criteria: (I) a case-control, nested case-control, or cohort design; (II) evaluate the rs712 polymorphism among various cancer patients and controls; (III) available genotype data for extraction to calculate ORs with 95% CIs; (IV) when duplications or the same patients used in several publications existed, we only chose the most recent or complete study in the meta-analysis. Additionally, the major exclusion criterion was as follows: (I) no available genotype frequency data; (II) non case-control studies, case reports, letters, reviewed editorial articles; (III) duplicated publications with previous studies.
Data extraction
Two researchers (B Yu and X Wang) inspected studies independently and extracted appropriate information and data with a standard protocol, to ensure the reliability and the accuracy of the results. Moreover, the results were reviewed by a third investigator. We extracted the following information form from each study: name of first author, publication date, type of carcinoma, ethnicity, source of controls, methods of genotyping, and total number and genotype frequencies (GG, GT or TT respectively) of cases and controls of rs712. Moreover, the Hardy-Weinberg equilibrium (HWE) test results were also recorded.
Statistical analysis
The following contrasts for rs712 polymorphism and cancer risk were evaluated, according to five genetic comparison models: homozygous model (GG vs. TT), heterozygous model (GT vs. TT), dominant model (GT + GG vs. TT), recessive model (GG vs. TT + GT) and allele model (G vs. T). The pooled ORs with 95% CIs were presented to assess the strength of association between rs712 and susceptibility to cancer. Based on the detection of heterogeneity, the fixed-effects model or the random-effects model was selected in the present meta-analysis. The χ2-based Q-statistic was utilized to assess the Between-study heterogeneity, and P<0.05 was deemed statistically significant. The ORs were pooled according to the fixed-effects model, if the result of the heterogeneity test was P>0.05. Moreover, we also tested whether genotype frequencies of controls were in HWE by the goodness-of-fit chi-square test and P<0.05 was regarded significant disequilibrium (14). Sensitivity analysis was performed by calculating the stability of results to assess the influence of a single study in this meta-analysis once at a time. Additionally, publication bias was investigated using Begg’s funnel plot, and funnel plot asymmetry was further assessed by the Egger linear regression test. When the P value of the Egger test was <0.05, statistical significance was considered (15). All statistical analysis was conducted by Stata software (version 12.0; StataCorp LP, College Station, TX, USA).
Trial sequential analysis
Because of limited data and repetitive calculating of accumulating data, cumulative meta-analyses may increase the risks of producing type I and type II errors (16-18). Thus, the trial sequential analysis software (TSA, version 0.9; Copenhagen Trial Unit, Copenhagen, Denmark, 2011) was applied to assess the risk of type I errors in the cumulative meta-analysis, which combined an estimation of required information size with an adjusted threshold for statistical significance with dispersed data (18,19). Besides, the trial sequential monitoring boundaries were used to decide whether a trial could be terminated early and adjusted the CIs and reduces type I errors (20). If the cumulative Z-curve exceeds the required information size or crosses the trial sequential monitoring boundary, the results suggest a sufficient level of evidence in this meta-analysis is reached and no further studies are required. Otherwise, the obtained results are insufficient (21). In the present meta-analysis, TSA was performed to calculate the required information size and construct the trial sequential monitoring boundaries, based on an overall type-I error of 5%, a relative risk reduction of 20% in mortality and overall morbidity and a statistical test power of 80%.
Results
Studies characteristics
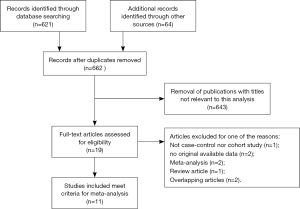
As shown in Figure 1, 662 records of rs712 and cancer were retrieved. After screening titles and abstracts of relevant articles, 643 articles were excluded because they were not related to the inclusion criteria. Furthermore, nineteen candidate studies were excluded after complete reading. Finally, eleven case-control studies were included in the meta-analysis, published between 2008 and 2015. All of them were retrospective in design.
The present meta-analysis including 3,572 cases of patients with various carcinomas and 4,749 controls from a total of eleven case-control studies on rs712 polymorphism and cancer risk (9,10,22-30), and the detailed data of each study is listed in Table 1. Besides, the distribution of genotypes in the controls of all studies was consistent with HWE. The flowchart of literature search and selection process is shown in Figure 1. The types of cancers in these studies included breast cancer (BC), CRC, cervical squamous cell carcinoma (CSCC), papillary thyroid cancer (PTC), nasopharyngeal carcinoma (NPC), glioma, NSCLC and GC (9-14). In these studies, three genotyping methods were applied, such as iMLDR, PCR-RFLP and AS-PCR to assay the polymorphism in rs712. Besides, ten studies were conducted on Asian population, and only one study was carried out on Caucasian population. Furthermore, we divided them into population-based group or hospital-based group in all studies, to distinguish between different sources of control group.
Table 1
| Year | Surname | Ethnicity | SOC | Genotyping | Cancer type | Case | Control | Case (n) | Control(n) | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GT | TT | GG | GT | TT | |||||||||||
| 2015 | Huang | Asian | HB | PCR-RFLP | BC | 228 | 251 | 155 | 65 | 8 | 173 | 71 | 7 | Yes | ||
| 2015 | Jiang | Asian | PB | PCR-RFLP | CRC | 586 | 476 | 372 | 176 | 38 | 331 | 133 | 12 | Yes | ||
| 2015 | Dai | Asian | PB | iMLDR | CRC | 430 | 430 | 253 | 145 | 32 | 283 | 130 | 17 | Yes | ||
| 2015 | Liang | Asian | HB | PCR-RFLP | CSCC | 415 | 504 | 257 | 144 | 14 | 327 | 163 | 14 | Yes | ||
| 2014 | Jin | Asian | HB | PCR-RFLP | PTC | 252 | 290 | 154 | 84 | 14 | 183 | 92 | 15 | Yes | ||
| 2014 | Pan | Asian | HB | PCR-RFLP | NPC | 188 | 356 | 112 | 64 | 12 | 201 | 138 | 17 | Yes | ||
| 2014 | Pan | Asian | HB | PCR-RFLP | CRC | 339 | 313 | 188 | 125 | 26 | 203 | 100 | 10 | Yes | ||
| 2013 | Li | Asian | HB | PCR-RFLP | GC | 181 | 674 | 105 | 60 | 16 | 442 | 211 | 21 | Yes | ||
| 2013 | Yan | Asian | HB | PCR-RFLP | Glioma | 153 | 204 | 83 | 56 | 14 | 137 | 61 | 6 | Yes | ||
| 2010 | Peng | Asian | HB | PCR-RFLP | NSCLC | 83 | 80 | 49 | 31 | 3 | 51 | 25 | 4 | Yes | ||
| 2008 | Landi | Caucasian | HB | AS-PCR | CRC | 717 | 1171 | 239 | 341 | 137 | 364 | 571 | 236 | Yes | ||
SOC, source of controls; PB, population-based controls; HB, hospital-based controls; BC, breast cancer; CRC, colorectal cancer; CSCC, cervical squamous cell carcinoma; HCC, hepatocellular carcinoma; PTC, papillary thyroid cancer; NPC, nasopharyngeal carcinoma; NSCLC, non-small cell carcinoma; GC, gastric cancer; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; AS-PCR, allele specific polymerase chain reaction; iMLDR, improved multi-plex ligation detection reaction.
3Quantitative synthesis results
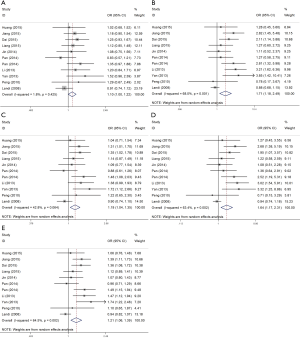
In this meta-analysis, the random-effect model was selected to calculate the pooled OR with corresponding 95% CIs except heterozygote model, because the between-study heterogeneity among those studies was obvious (P<0.05). The combined results showed that rs712 polymorphism was significantly associated with cancer risk. Overall, the main results of this meta-analysis about the associations between rs712 polymorphism and the risk of cancer were listed in Table 2. The meta-analytic results showed that significant cancer risk was associated with rs712 for heterozygote model OR =1.10 (95% CI: 1.002–1.22), homozygote model OR =1.71 (95% CI: 1.18–2.49), dominant model OR =1.19 (95% CI: 1.04–1.35), recessive model OR =1.64 (95% CI: 1.17–2.31) and allele model OR =1.21 (95% CI: 1.06–1.39) (Figure 2). Moreover, for the first time, TSA were performed to confirm such associations, demonstrating that the findings in the current study were proved to be solid with sufficient evidence. In total, with the effect of rs712 gene polymorphism, the carriers of T allele held higher cancer risk than carriers of G allele.
Table 2
| Na | Sample size | GT vs. GG | TT vs. GG | GT/TT vs. GG | TT vs. GT/GG | T vs. G | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)* | Pb | OR (95% CI)* | Pb | OR (95% CI)* | Pb | OR (95% CI)* | Pb | OR (95% CI)* | Pb | ||||||
| 11 | 8,321 | 1.10 (1.002–1.22) | 0.425 | 1.71 (1.18–2.49) | 0.001 | 1.19 (1.04–1.35) | 0.064 | 1.64 (1.17–2.31) | 0.002 | 1.21 (1.06–1.39) | 0.002 | ||||
a, number of studies; b, P value of Q test for heterogeneity; *, random-effects model was used when P value for heterogeneity test <0.1; otherwise, fixed-effects model was used.
Test of heterogeneity
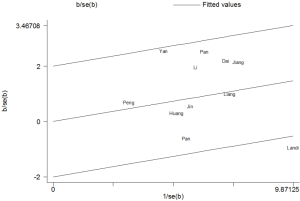
Because of the heterogeneity, a Galbraith radial plot was utilized under the dominant model (Figure 3), which also indicated that no significant heterogeneity between studies was observed.
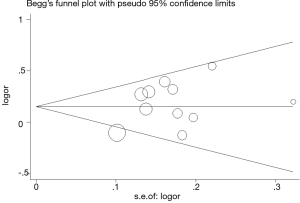
Publication bias
The potential publication bias for the all available data was assessed by the Begg’s funnel plot and Egger’s test. The shape of the funnel plot seemed symmetrical in the funnel plot analysis (Figure 4). No publication bias was demonstrated in the Begg’s test (P=0.679) and Egger’s test in the dominant model (P=0.584).
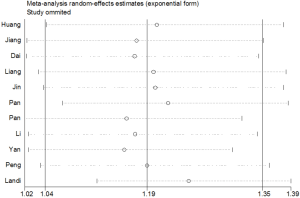
Sensitivity analysis
Sensitivity analysis was performed to evaluate the influence of each study on the pooled OR by omitting each individual studies in turn. Figure 5 showed the sensitivity analysis under the dominant model of the association of rs712 and cancer risk, demonstrating that our results were reliable and robust.
Trial sequential analysis results
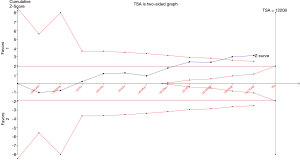
In our current study, the cumulative Z-curve exceeded the trial sequential monitoring boundary (Figure 6), suggesting sufficient evidence of the association between rs712 polymorphism and risk of cancer.
Discussion
In recent years, increasing studies in cancer research focused on the field of miRNAs, which form an important class of regulators and participate in various functions including apoptosis, development and cell proliferation (30,31). Considering miRNAs play important roles in the pathogenesis of cancer, it might be regarded as potential biomarkers for cancer risk or development. The KRAS gene was confirmed to promote tumorigenesis through the pathway of RAF/MEK/MAPK (32). KRAS mutation with a single amino acid substitution could lead to an activating mutation or increasing KRAS expression, and might have connections with cancers. Let-7 could bind to specific site in 3′UTR of KRAS mRNA so as to down-regulate RAS and then function as tumor-suppressing gene (8). Additionally, rs712, as one of the 8–10 putative let-7 miRNA complementary sites, might adjust KRAS expression (5).
Recently, a growing number of studies involved the relationship between rs712 and risk of various carcinomas, but the results were inconclusive. Li et al. found that there was significant difference in the TT genotype of rs712 polymorphism between the GC patients and the controls group, when GG genotyping was taken as a reference (OR =3.05, 95% CI: 1.53–6.08). Thus, we suggested that the KRAS rs712 polymorphism might be a genetic marker in the development of GC (9). In addition, Pan et al. showed that the genotype and allele frequencies of the rs712 polymorphism was associated with an increased risk of NPC, compared to the controls group (GT vs. GG, OR =0.83, 95% CI: 0.51–1.21; TT vs. GG, OR =1.27, 95% CI: 0.58–2.75) (28). However, studies from Wang et al. made the opposite results, which showed that GT or TT genotyping were protecting factors for OSCC (OR =0.26, 95% CI: 0.10–0.60) (11). Thus, we conducted this meta-analysis to investigate the relationship between rs712 polymorphism and cancer risk. The results of this meta-analysis indicated that G>T variant of rs712 increase cancer risk mainly in Asians.
Compared to a single study especially in analyzing unexplained studies, meta-analysis is a powerful tool and can provide more sufficient results (33). As a result, we suggested there existed a much stronger advantage to prove the association between rs712 with cancer susceptibility. The present meta-analysis deemed that the let-7 genetic polymorphism might influence the risk of cancer. In these previous studies, these outcomes remained unclear. Therefore, we need a better method to analyze and understand the association between let-7 polymorphism and susceptibility to cancer. In addition, for the first time, TSA was adopted to reduce the risk of type I error and testify whether our results were established on firm evidence of effect. In the current meta-analysis, our results revealed the T allele of let-7 genetic polymorphism rs712 increases cancer susceptibility, mainly among Asian ethnicity.
As a powerful and useful approach, TSA is introduced to calculate the required information size for the meta-analysis with the adaptation of monitoring boundaries and to reduce the risk of type I error and estimate whether further trials are needed (20). Compare to the traditional meta-analysis, more reliable evidence is obtained by TSA. Here, when an additional study was involved, an updated meta-analysis was performed and the cumulative Z-curve of each meta-analysis was constructed, so as to dictate whether a sufficient level of evidence has been reached or the futility boundary is reached (21). In the present meta-analysis, the cumulative Z-curve exceeded the trial sequential monitoring boundary, which meant that our results were established on firm evidence of effect.
Overall, sufficient statistical evidence including the large sample size and the implementation of TSA were used to estimate such association. However, there are several limitations were involved in our study. Firstly, the studies included in this meta-analysis were mainly conducted on Asians and only a study was Caucasian populations. Besides, there are no reports on the other population available. Thus, more researches should pay attention to the influence of ethnicity factors in the future. Secondly, only eleven studies were included in this study, which might weaken the reliability of our meta-analysis. Therefore, more well-designed studies with large cases of each specific cancer should be conducted to validate the relationship between let-7 polymorphism rs712 and cancer risk. Besides, the results of this meta-analysis were that based on unadjusted estimates, so we should take into consideration the effect of multiple confounders such as life-style, age, environmental factors and so on. Moreover, the obvious heterogeneity existed in our study probably due to the difference in the tumor types. Hence, to improve reliability of the meta-analysis, more studies should need to investigate rs712 and risk of cancer in the subsequent years.
Conclusions
This meta-analysis suggested that a let-7 binding site polymorphism rs712 in the KRAS 3′UTR increased the risk of cancer in Asians and it has the potential to be a prognostic factor in various carcinoma in the future. To further evaluate the relationship between genetic polymorphism rs712 and susceptibility for every specific carcinoma, large and better-designed studies are needed.
Acknowledgements
Funding: None.
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2017.05.07). Dr. Xiao Li serves as an unpaid section editor of AME Medical Journal. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This article does not contain any studies with human participants or animals performed by any of the authors.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol 2011;3:140-55. [PubMed]
- Liu J, Zheng M, Tang YL, et al. MicroRNAs, an active and versatile group in cancers. Int J Oral Sci 2011;3:165-75. [Crossref] [PubMed]
- Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol 2011;38:724-33. [Crossref] [PubMed]
- Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta 2005;1756:81-2.
- Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res 2008;68:8535-40. [Crossref] [PubMed]
- Saridaki Z, Weidhaas JB, Lenz HJ, et al. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clin Cancer Res 2014;20:4499-510. [Crossref] [PubMed]
- Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 2009;10:399-416. [Crossref] [PubMed]
- Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. [Crossref] [PubMed]
- Li ZH, Pan XM, Han BW, et al. A let-7 binding site polymorphism rs712 in the KRAS 3' UTR is associated with an increased risk of gastric cancer. Tumour Biol 2013;34:3159-63. [Crossref] [PubMed]
- Pan XM, Sun RF, Li ZH, et al. A let-7 KRAS rs712 polymorphism increases colorectal cancer risk. Tumour Biol 2014;35:831-5. [Crossref] [PubMed]
- Wang WY, Chien YC, Wong YK, et al. Effects of KRAS mutation and polymorphism on the risk and prognosis of oral squamous cell carcinoma. Head Neck 2012;34:663-6. [Crossref] [PubMed]
- Ying HQ, Wang F, He BS, et al. The involvement of Kras gene 3'-UTR polymorphisms in risk of cancer and influence on patient response to anti-EGFR therapy in metastatic colorectal cancer: a meta-analysis. Onco Targets Ther 2014;7:1487-96. [PubMed]
- Zhao WH, Qu XF, Xing ZG, et al. Association of rs712 polymorphism in Kras gene 3'-luntranslated region and cancer risk: a meta-analysis. J BUON 2015;20:309-16. [PubMed]
- Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992;48:361-72. [Crossref] [PubMed]
- Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiol 2005;15:235-43. [Crossref] [PubMed]
- Brok J, Thorlund K, Wetterslev J, et al. Apparently conclusive meta-analyses may be inconclusive--Trial sequential analysis adjustment of random error risk due to repetitive testing of accumulating data in apparently conclusive neonatal meta-analyses. Int J Epidemiol 2009;38:287-98. [Crossref] [PubMed]
- Turner RM, Bird SM, Higgins JP. The impact of study size on meta-analyses: examination of underpowered studies in Cochrane reviews. PLoS One 2013;8:e59202. [Crossref] [PubMed]
- Wetterslev J, Thorlund K, Brok J, et al. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J Clin Epidemiol 2008;61:64-75. [Crossref] [PubMed]
- Bangalore S, Kumar S, Wetterslev J, et al. Angiotensin receptor blockers and risk of myocardial infarction: meta-analyses and trial sequential analyses of 147 020 patients from randomised trials. BMJ 2011;342:d2234. [Crossref] [PubMed]
- Thorlund K, Devereaux PJ, Wetterslev J, et al. Can trial sequential monitoring boundaries reduce spurious inferences from meta-analyses? Int J Epidemiol 2009;38:276-86. [Crossref] [PubMed]
- Holst LB, Petersen MW, Haase N, et al. Restrictive versus liberal transfusion strategy for red blood cell transfusion: systematic review of randomised trials with meta-analysis and trial sequential analysis. BMJ 2015;350:h1354. [Crossref] [PubMed]
- Dai Q, Wei HL, Huang J, et al. KRAS polymorphisms are associated with survival of CRC in Chinese population. Tumour Biol 2016;37:4727-34. [Crossref] [PubMed]
- Huang X, Yang Y, Guo Y, et al. Association of a let-7 KRAS rs712 polymorphism with the risk of breast cancer. Genet Mol Res 2015;14:16913-20. [Crossref] [PubMed]
- Jiang QH, Peng HX, Zhang Y, et al. rs712 polymorphism within let-7 microRNA-binding site might be involved in the initiation and progression of colorectal cancer in Chinese population. Onco Targets Ther 2015;8:3041-5. [PubMed]
- Jin H, Liang Y, Wang X, et al. Association between a functional polymorphism rs712 within let-7-binding site and risk of papillary thyroid cancer. Med Oncol 2014;31:221. [Crossref] [PubMed]
- Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 2008;29:579-84. [Crossref] [PubMed]
- Liang Y, Sun R, Li L, et al. A Functional Polymorphism in the Promoter of MiR-143/145 Is Associated With the Risk of Cervical Squamous Cell Carcinoma in Chinese Women: A Case-Control Study. Medicine (Baltimore) 2015;94:e1289. [Crossref] [PubMed]
- Pan XM, Jia J, Guo XM, et al. Lack of association between let-7 binding site polymorphism rs712 and risk of nasopharyngeal carcinoma. Fam Cancer 2014;13:93-7. [Crossref] [PubMed]
- Yan L, Wang Q, Tian T. Association between the single nucleotide polymorphism of let-7 target gene KRAS-binding site rs712 and risk of glioma. Chinese Journal of Cancer Prevention and Treatment 2013;20:811-4.
- Peng X, Zhao J, Lei Z, et al. Association of A SNP in MicroRNA Let-7 Complementary Region in KRAS 3’UTR with Non-small Cell Lung Cancer. Journal of Soochow University (Medical Science Edition) 2010;786-90.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- van Grieken NC, Aoyama T, Chambers PA, et al. KRAS and BRAF mutations are rare and related to DNA mismatch repair deficiency in gastric cancer from the East and the West: results from a large international multicentre study. Br J Cancer 2013;108:1495-501. [Crossref] [PubMed]
- Munafò MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry Res 2004;129:39-44. [Crossref] [PubMed]
Cite this article as: Yu B, Wang X, Qin Z, Xue J, Cai H, Zhang C, Xu W, Wang J, Li X, Xu Z, Xu T, Zou Q. The association between let-7 microRNA-binding site polymorphism rs712 and cancer risk: a meta-analysis and trial sequential analysis. AME Med J 2017;2:59.