
Circulating cell-free DNA is a potential prognostic biomarker of metastatic castration-resistant prostate cancer for taxane therapy
Prostate cancer (PC) is the most frequently diagnosed cancer and a major cause of cancer deaths among men in the developed countries (1). Metastatic prostate cancer remains the major challenge in PC treatment. Despite standard treatment with androgen deprivation therapy (ADT), patients with metastatic PC will inevitably progress to metastatic castration-resistant prostate cancer (mCRPC). In recent years, a number of therapeutic options for mCRPC have been developed, including taxane-based chemotherapy and androgen receptor (AR)-targeting therapy with either abiraterone or enzalutamide (2,3). Although not curative, these treatments prolong patient survival. In this regard, the selection of appropriate treatment plan to maximize survival benefit becomes important. Of note, extensive research effort has been devoted in exploring prognostic biomarkers to assess disease progression under second line therapies in men with mCRPC. Using the CALGB-90401 phase III clinical trial (n=1,050) and validation with a subgroup (n=942) in the ENTHUSE 33 trial, a prognostic model was recently updated to assess overall survival (OS) in mCRPC patients receiving the first line chemotherapy with docetaxel. The model consists of eight baseline clinical factors: opioid analgesic use, lactate dehydrogenase (LDH), disease site, Eastern Cooperative Oncology Group (ECOG) performance status, albumin, hemoglobin, alkaline phosphatase (ALP), and prostate-specific antigen (PSA) (4). Recently, an even more powerful risk estimate model, the ensemble of penalized Cox regression (ePCR) model, has been reported to predict OS in mCRPC patients receiving docetaxel treatment. This model was generated by a massive collaborative effort through the Dialogue for Reverse Engineering Assessments and Methods (DREAM) challenge platform and involved five phase III clinical trials (n=2,336). In addition to the parameters described above, this model also include many other factors related to kidney function, haematology, and others (5,6).
In an effort to develop additional prognostic biomarkers of mCRPC for taxane-based chemotherapy, Mehra et al. performed a post-hoc analysis of two phase III clinical trials to estimate the biomarker values of plasma cell-free DNA (pcfDNA) for disease progression on taxane-based chemotherapy (7). Chemotherapy-naïve patients with mCRPC in the FIRSTANA trial were receiving either cabazitaxel or docetaxel (8); post-docetaxel mCRPC patients in the PROSELICA trial were treated with cabazitaxel (9). Drugs were administered intravenously once every three weeks (Q3W) for multiple cycles; blood was collected prior to initiating cycles 1, 2, 4, and at the end of therapy (Figure 1). A total of 571 patients with 315 from FIRSTANA (n=1,168) and 256 from PROSELICA (n=1,200) had blood collected and analyzed in this study (7). Cell-free DNA in plasma was subsequently isolated and quantified. The pcfDNA concentrations from baseline, cycle 2 (C2), and C4 were analyzed for associations with PSA response using logistic regression, radiological progression free survival (rPFS), and OS using Cox models (Figure 1). Patients from individual trials were analyzed separately and in combination; in the latter setting, a two-stage individual patient meta-analysis was performed and heterogeneity was controlled with I2 statistics (7).
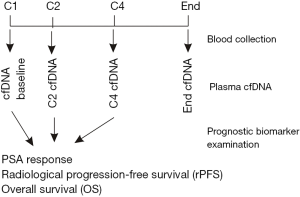
Baseline pcfDNA concentration was not associated with PSA response (defined by the Prostate Cancer Working Group 2 as ≥50% of PSA reductions) and radiological response in FIRSTANA, PROSELICA, and the combination populations (Table 1) (7). Nonetheless, longitudinal pcfDNA concentrations were significantly lower per cycle treatment in patients with PSA response in comparison to those without PSA response (7). Additionally, C2 and C4 pcfDNA concentrations were significantly associated with PSA response in FIRSTANA, PROSELICA, and combined patient populations (Table 1) (7). Collectively, baseline pcfDNA concentration does not have a clear predictive value for PSA response in patients with mCRPC receiving chemotherapy, while pcfDNA concentrations during treatment are predictive of PSA response. However, it should be noticed that the reduction of pcfDNA concentration were only transiently observed in patients with PSA response in cycles 2 and 4 treatment and that the C2 concentration of pcfDNA did not associate with PSA response in FIRSTANA patients (Table 1) (7).
Table 1
| pcfDNA | Outcome | Study patients | Association status |
|---|---|---|---|
| Baseline | PSA responsei and radiological responsei | FIRSTANA | No |
| PROSELICA | No | ||
| Combined | No | ||
| rPFSii | FIRSTANA | Yes (HR =1.82/95% CI, 1.13–2.95) | |
| PROSELICA | No (1.26/0.72–2.20) | ||
| Combined | Yes (1.56/1.08–2.95) | ||
| OSii | FIRSTANA | Yes (1.80/1.16–2.79) | |
| PROSELICA | No (1.36/0.91–2.05) | ||
| Combined | Yes (1.55/1.15–2.09) | ||
| Longitudinal | PSA responsei | FIRSTANA | No (C2 pcfDNA), yes (C4 pcfDNA) |
| C2 | PROSELICA | Yes (C2 pcfDNA), yes (C4 pcfDNA) | |
| C4 | Combined | Yes (C2 pcfDNA), yes (C4 pcfDNA) | |
| rPFSii | Combined | Yes (1.89/1.36–2.63, P<0.001) (C2 pcfDNA) | |
| Yes (1.88/1.32–2.68, P<0.001) (C4 pcfDNA) | |||
| OSii | Combined | Yes (1.77/1.37–2.29, P<0.001) (C2 pcfDNA) | |
| Yes (1.75/1.30–2.35, P<0.001) (C4 pcfDNA) |
i, determined by univariable logistic regression; ii, modeled through multivariable Cox analysis.
Unlike PSA response, baseline pcfDNA concentration was significantly associated with rPFS with adjusted hazard ratio (aHR 1.54), 95% confidence interval (95% CI, 1.15–2.08), and P=0.004; and OS (aHR 1.53, 95% CI, 1.18–1.97, P=0.001) in the combined population (7). Multivariable Cox model of baseline pcfDNA concentration together with other baseline characteristics including ECOG performance status, visceral metastasis, bone-only disease, Gleason score, pain, albumin, ALP, LDH, and NLR (neutrophil-to-lymphocyte ratio) revealed the baseline concentration of pcfDNA being an independent prognostic factor for rPFS and OS in the combined population (Table 1) (7). Both C2 and C4 pcfDNA concentrations also predicted rPFS (HR 1.89, 95% CI, 1.36–2.63, P<0.001; HR 1.88, 95% CI, 1.32–2.68, P<0.001, respectively) and OS (HR 1.77, 95% CI, 1.37–2.29, P<0.001; HR 1.75, 95% CI, 1.30–2.35, P<0.001, respectively) (Table 1) (7). For analysis of individual patient populations, baseline pcfDNA concentration associates with rPFS in FIRSTANA (HR 1.82, 95% CI, 1.13–2.95) but not PROSELICA (HR 1.26, 95% CI, 0.72–2.20); and OS in FIRSTANA (HR 1.80, 95% CI, 1.16–2.79) but not PROSELICA (HR 1.36, 95% CI, 0.91–2.09) (Table 1) (7). Furthermore, using C-index, the inclusion of baseline pcfDNA did not improve the fit of the multivariable model consisting of all aforementioned baseline characteristics in discriminating either rPFS or OS when pcfDN was not included (7). These observations suggest that pcfDNA concentration may not be a robust classifier for response to taxane chemotherapy.
Nonetheless, the study by Mehra et al. demonstrated an overall utility of pcfDNA quantitation in predicting mCRPC progression on taxane-based chemotherapy. In both FIRSTANA and PROSELICA groups, baseline pcfDNA concentration was significantly associated with a set of prognostic characteristics including ECOG performance status, pain at baseline, albumin, ALP, haemoglobin, LDH, PSA doubling time (<2 vs. ≥2 months), and NLR at baseline (7). These associations are in accordance with the reported cumulative evidence for a general association of circulating cfDNA (ccfDNA) with PC tumorigenesis and progression (10). Elevations of ccfDNA concentration were reported in PC patients compared to individuals with benign prostatic hyperplasia (BPH; patients n=142 vs. BPH n=19) (11) and healthy controls (patients n=133 vs. controls n=33) (12). High levels of baseline ccfDNA were observed to associate with biochemical recurrence (BCR) in patients with localized disease subjected to radical prostatectomy (13). Although the entry concentrations of ccfDNA did not correlate with BCR, an increase in ccfDNA following surgery was associated with a reduction in BCR free survival (12). Among the 48 mCRPC patients with PSA declines following taxane-based chemotherapy, patients (n=6) with PSA reductions less than 30% displayed a significantly higher baseline ccfDNA compared to others; however, baseline ccfDNA concentration was not associated with PSA declines ≥50% (14), which was consistent with the observations reported by Mehra et al. (7). Collectively, it does not appear that baseline ccfDNA quantitation has a prognostic value for PSA response (reductions ≥50%) in mCRPC patients treated with taxane-based chemotherapy.
In the combined study and FIRSTANA population of docetaxel-naïve patients with mCRPC, baseline pcfDNA concentrations were significantly associated with shortening in rPFS and OS (Table 1) (7). The associations did not reach statistical significance in the PROSELICA population of post-docetaxel patients judged on the low boundary of 95% CI less than 1 (Table 1) (7). As this result was derived from multivariable Cox analyses (7); it is possible that univariable Cox analysis might reveal baseline pcfDNA concentration being a significant risk factor of reductions in rPFS and OS in the PROSELICA population following second line cabazitaxel chemotherapy. Nonetheless, evidence suggests that the prognostic value of baseline pcfDNA concentration for rPFS and OS seems to be reduced in mCRPC patients progressed on docetaxel therapy. The inclusion of baseline pcfDNA concentration did not enhance the fit of the model composed of the clinical characteristics with demonstrated prognostic value for OS in mCRPC patients treated with chemotherapy (4,5,7). In this situation, the prognostic value of baseline pcfDNA concentration for rPFS and OS would be more appreciated should the C-index of baseline pcfDNA alone in the separation of radiological progression and fatality was reported (7).
The presence of ccfDNA in healthy individuals was reported 70 year ago (15); its levels are elevated in response to a variety of pathological conditions including benign lesions, cancer, rheumatoid arthritis, inflammation, and tissue trauma (16-18). Quantitation of ccfDNA is thus not cancer-specific, which likely contributes to the limitation of using baseline pcfDNA concentration to predict mCRPC progression on taxane-based chemotherapy (7).
Unlike quantitative alterations, qualitative changes in ccfDNA mirror tumor-associated genomic alterations. With next generation sequencing (NGS), detection of genome instability can impressively discriminate PC from non-tumor tissue with an area under curve (AUC) of 0.92 (95% CI, 0.87–0.95) (19). Methylation of CpG islands was detected in serum cfDNA, which displayed a diagnostic value on PC (20). Consistent with persistent AR signaling being a major cause in disease progression on AR-targeting therapies (21), copy number increases in the AR and CYP17A1 (a critical enzyme functioning in androgen biosynthesis) genes observed in ccfDNA significantly associate with disease progression following abiraterone, enzalutamide, and taxane therapies (22,23). Furthermore, NGS-based deep exon sequencing of 72 mCRPC driver genes using pcfDNA detected somatic mutations in 72 genes with a high level of concordance with tumor associated mutations (R2 =0.9) in treatment-naïve mCRPCs to second line AR-targeted therapy of abiraterone or enzalutamide (24). Among these mutations, defects in BRCA2 and ATM strongly predict poor outcome (24). In this regard, there exists great opportunities for future studies to investigate the prognostic value of AR abnormalities, genomic alterations in the 72 mCRPC driver genes, and epigenetic alterations in ccfDNA for rPFS and OS in patients with mCRPC to taxane-based chemotherapy using FIRSTANA and PROSELICA clinical trials.
Qualitative analyses of ccfDNA using comprehensive clinical materials available in phase III trials will certainly enhance its ability to predict treatment response in patients with mCRPC and improve clinical decision making regarding treatment regimen choices. This will make the non-invasive ccfDNA-based liquid biopsy even more attractive. However, ccfDNA as a biomarker is not without its limitations. Although evidence suggests apoptosis and necrosis as mechanisms response for releasing cellular DNA into circulation, there is a lack of comprehensive understanding on the process of how blood cfDNA are produced (25). Without detailed knowledge, it is difficult to envisage how the ccfDNA composition reflects tumor-associated heterogeneity. For example, prostate cancer stem cells may be quiescent and their DNA content will unlikely be released through cell damaging mechanisms including apoptosis and necrosis. A large body of evidence exists to support a critical role of prostate cancer stem cells in PC progression under ADT and second line therapies involving abiraterone, enzalutamide, and taxane (21). Furthermore, blood cfDNA is under active clearance by the liver and kidney (25), which results their presence at low levels. This clearance may also alter the composition of blood cfDNA, thereby compromising its representation of tumor-associated heterogeneity.
A number of treatment options are available to patients with mCRPC, none are curative. Good biomarkers or prognostic models facilitate decision making in treatment selection. Collective effort in this domain has indeed formulated a set of clinical characters to predict outcome in patients with mCRPC, which include bone pain, LDH, disease site, ECOG performance status, albumin, hemoglobin, ALP, PSA, and others (4,5). Further research in the field of biomarker exploration for mCRPC should include mechanism-based molecular events in addition to those of demonstrated clinical characteristics.
Acknowledgements
Funding: D Tang is supported by an Award from Teresa Cascioli Charitable Foundation Research Award in Women’s Health and grants from Canadian Cancer Society (grant#: 319412) and Cancer Research Society.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xiao Li (Department of Urologic Surgery, the Affiliated Cancer Hospital of Jiangsu Province of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2018.06.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364:1995-2005. [Crossref] [PubMed]
- Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187-97. [Crossref] [PubMed]
- Halabi S, Lin CY, Kelly WK, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol 2014;32:671-7. [Crossref] [PubMed]
- Guinney J, Wang T, Laajala TD, et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol 2017;18:132-42. [Crossref] [PubMed]
- Mei W, Kapoor A, Major P, et al. Progress towards accurate prediction of overall survival in men with metastatic castration-resistant prostate cancer. Journal of Xiangya Medicine 2017;2:17. [Crossref]
- Mehra N, Dolling D, Sumanasuriya S, et al. Plasma Cell-free DNA Concentration and Outcomes from Taxane Therapy in Metastatic Castration-resistant Prostate Cancer from Two Phase III Trials (FIRSTANA and PROSELICA). Eur Urol 2018; [Epub ahead of print]. [Crossref]
- Oudard S, Fizazi K, Sengelov L, et al. Cabazitaxel Versus Docetaxel As First-Line Therapy for Patients With Metastatic Castration-Resistant Prostate Cancer: A Randomized Phase III Trial-FIRSTANA. J Clin Oncol 2017;35:3189-97. [Crossref] [PubMed]
- Eisenberger M, Hardy-Bessard AC, Kim CS, et al. Phase III Study Comparing a Reduced Dose of Cabazitaxel (20 mg/m(2)) and the Currently Approved Dose (25 mg/m(2)) in Postdocetaxel Patients With Metastatic Castration-Resistant Prostate Cancer-PROSELICA. J Clin Oncol 2017;35:3198-206. [Crossref] [PubMed]
- Yin C, Luo C, Hu W, et al. Quantitative and Qualitative Analysis of Circulating Cell-Free DNA Can Be Used as an Adjuvant Tool for Prostate Cancer Screening: A Meta-Analysis. Dis Markers 2016;2016:3825819.
- Chun FK, Muller I, Lange I, et al. Circulating tumour-associated plasma DNA represents an independent and informative predictor of prostate cancer. BJU Int 2006;98:544-8. [Crossref] [PubMed]
- Wroclawski ML, Serpa-Neto A, Fonseca FL, et al. Cell-free plasma DNA as biochemical biomarker for the diagnosis and follow-up of prostate cancer patients. Tumour Biol 2013;34:2921-7. [Crossref] [PubMed]
- Bastian PJ, Palapattu GS, Yegnasubramanian S, et al. Prognostic value of preoperative serum cell-free circulating DNA in men with prostate cancer undergoing radical prostatectomy. Clin Cancer Res 2007;13:5361-7. [Crossref] [PubMed]
- Kienel A, Porres D, Heidenreich A, et al. cfDNA as a Prognostic Marker of Response to Taxane Based Chemotherapy in Patients with Prostate Cancer. J Urol 2015;194:966-71. [Crossref] [PubMed]
- Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Leon SA, Ehrlich GE, Shapiro B, et al. Free DNA in the serum of rheumatoid arthritis patients. J Rheumatol 1977;4:139-43. [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta 2007;1775:181-232.
- Schutz E, Akbari MR, Beck J, et al. Chromosomal instability in cell-free DNA is a serum biomarker for prostate cancer. Clin Chem 2015;61:239-48. [Crossref] [PubMed]
- Ellinger J, Haan K, Heukamp LC, et al. CpG island hypermethylation in cell-free serum DNA identifies patients with localized prostate cancer. Prostate 2008;68:42-9. [Crossref] [PubMed]
- Ojo D, Lin X, Wong N, et al. Prostate Cancer Stem-like Cells Contribute to the Development of Castration-Resistant Prostate Cancer. Cancers (Basel) 2015;7:2290-308. [Crossref] [PubMed]
- Buelens S, Claeys T, Dhondt B, et al. Prognostic and Therapeutic Implications of Circulating Androgen Receptor Gene Copy Number in Prostate Cancer Patients Using Droplet Digital Polymerase Chain Reaction. Clin Genitourin Cancer 2018;16:197-205.e5. [Crossref] [PubMed]
- Salvi S, Casadio V, Conteduca V, et al. Circulating cell-free AR and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br J Cancer 2015;112:1717-24. [Crossref] [PubMed]
- Annala M, Vandekerkhove G, Khalaf D, et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov 2018;8:444-57. [Crossref] [PubMed]
- Volckmar AL, Sultmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018;57:123-39. [Crossref] [PubMed]
Cite this article as: Mei W, Gu Y, Jiang Y, Major P, Tang D. Circulating cell-free DNA is a potential prognostic biomarker of metastatic castration-resistant prostate cancer for taxane therapy. AME Med J 2018;3:68.

