
Circles in action, circles in function
Circular RNAs were identified more than 20 years ago and thought, at the time, to simply represent rare biochemical curiosities, indicative of an “error-prone” splicing machinery (1-3). It was not until recently, with the advent of next generation sequencing, that such non-linear transcripts were found to be ubiquitously expressed and in certain cases being more abundant that the corresponding linear transcripts (4). Moreover, functional properties have started to be assigned to these molecules, including binding to proteins and acting as miRNAs traps (5,6). In their recent work Yu et al provide convincing evidence for the functionality of a circular RNA from the SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5 (SMARCA5) locus, cSMARCA5, containing exons 15 and 16 of the SMARCA5 gene back-spliced together (7).
Using elegant and extensive analysis, the authors initially identified, from RNAseq data of five hepatocellular carcinoma (HCC) samples and the corresponding adjacent noncancerous liver (ANL), 4,142 circular RNAs originating from protein coding exons of 2,322 genes, 190 of which are differentially expressed in the HCC versus the ANL samples. Integrating expression data on the corresponding mRNAs revealed that about 30% of these circular RNAs are deregulated in the opposite direction compared to their linear transcripts in the HCC and ANL samples. This is indicative of functionality, since simple by-products of non-optimized splicing processes would be expected to accumulate in the same direction as the corresponding mRNA.
The authors focus on cSMARCA5, the circular RNA that is on top of their list of abundant circular RNAs expressed differentially in HCC/ANL and in the opposite direction than the corresponding mRNA. Its expression is reduced in HCC compared to ANL. Worth noting is that the abundance of this circular RNAs is, in fact, higher than the linear mRNA transcript (8), another indication of possible functionality. First they demonstrate that the presence of complementary, non-Alu sequences in the flanking introns promotes circularization of the SMARCA5 exons 15 and 16. Then that the DExH-Box Helicase 9 (DHX9), but not the additional RNA binding proteins adenosine deaminase 1 acting on RNA (ADAR1) and quaking (QKI), downregulates cSMARCA5 in HCC cell lines. Importantly, DHX9 expression is increased in HCC and negatively correlates to cSMARC5, providing a mechanistic rationalization to the reduced cSMARC5 expression. Also worth noting is that in mammary epithelial cells QK1 upregulates cSMARCA5 (9), again arguing that the production of this circular RNA is a result of context-specific regulatory events.
Subsequently, in a cohort of 163 human HCC and the corresponding ANL samples the authors demonstrate that low cSMARCA5 expression correlates with worse overall and recurrence-free survival. Biochemical analysis in HCC cell lines confirmed that modulating cSMARCA5 levels impacted on cell growth and migration, with low expression increasing and high expression decreasing these cellular outcomes. Moreover, subcutaneously or orthotopically implanted high cSMARCA5 expressing HCC cells reduced tumor growth compared to control cells. Similarly, in vivo injection of high cSMARCA5 expressing HCC cells in the tail vein of nude mice reduced lung metastasis relative to control cells. Thus, all data pinpoint to tumor suppressive properties of cSMARCA5 in HCC.
Circular RNAs and especially abundant circular RNAs are known to have the capacity to act as miRNA traps. The authors used standard miRNA prediction tools to identify potential miRNAs interacting with cSMARCA5 and selected 29, which are also expressed in HCC or ANL. Subsequently, they used the novel technique of circular RNA in vivo precipitation (circRIP) (10) to identify miRNAs from the 29-list that interact with cSMARCA5 in HCC cells. This analysis highlighted miR-17-3p and miR-181b-5p associating with cSMARCA5. Mutations of the two miRNA binding sites on cSMARCA5 followed by luciferase analysis confirmed these interactions, as also did the enrichment of cSMARCA5 by miR-17-3p and miR-181b-5p pull-down assays. Worth noting is that the two miRNA binding sites on cSMARCA5 are not positioned at the back-spliced junction of exon 16 to exon 15 but at the canonical splice junction of exon 15 to exon 16, in fact, the two binding sites partially overlap. This raises the possibility that linear SMARCA5 transcripts encompassing exon 15 and exon 16 could also be targeted by the two miRNAs.
miR-17-3p and miR-181b-5p are thought to promote HCC (11), consequently the authors focused on HCC tumor suppressors that can be targeted by the two miRNAs. They demonstrate that the tumor suppressor tissue inhibitor of metalloproteinase 3 (TIMP3), a target of both miRNAs, is highly regulated in HCC cells by changes in the expression of cSMARCA5. Consistent with a capacity of the circular RNA to trap miRNAs, high cSMARCA5 levels upregulate and low levels downregulate TIMP3. Moreover, and similar to cSMARCA5, TIMP3 expression was reduced in HCC, positively correlating with cSMARCA5 expression.
Thus, a mechanistic scenario consistent with a protective role of a circular RNA in HCC, which involves trapping miRNAs that downregulate a tumor suppressor, has been put forward.
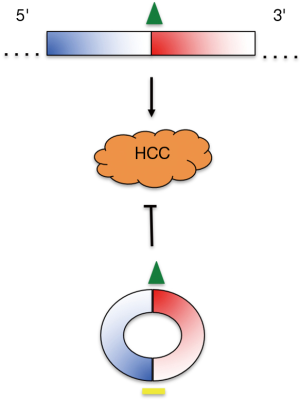
It should be noted that in HCC, and in contrast to the reduced cSMARCA5 levels, the SMARCA5 mRNA (and the SMARCA5 protein) is increased relative to ANL. Moreover, the observed SMARCA5 mRNA and protein upregulation is thought to promote HCC (12). Given that the miRNA target sites identified on exons 15 and 16 are present not only in cSMARCA5 but also in the corresponding linear transcript, then the regulatory scenarios may, in fact, be even more complicated. MiR-17-3p and miR-181b-5p targeting of the SMARCA5 mRNA could potentially destabilize it inhibiting HCC. Additionally, trapping the miRNAs on the linear transcript may reduce their availability for TIMP3 downregulation, also potentially inhibiting HCC. Consequently, not only the levels of cSMARCA5 but also the relative expression of circular to linear SMARCA5 RNAs encompassing exons 15 and 16, may also impact on HCC development and prognosis (Figure 1).
Is cSMARC5 the only circular RNA that affects HCC? Well, the authors have produced a substantial list of circular RNAs, which are expressed at reasonable levels and differentially regulated in HCC versus ANL. Some may simply be passengers, but others may indeed influence disease development.
Is miRNA trapping the only mechanism of action of circular RNAs in HCC? Well, testing for miRNA interactions is an established experimental approach, while the analysis of circular RNA interactions with proteins or other RNAs is not streamlined to the same extent, and consequently more difficult to address.
Taken together, the available evidence suggests that the potential of novel findings involving circular RNAs is, apparently, at hand and applying logical criteria for optimized selections, combined with technical innovations, will certainly pinpoint to additional functional implications of non-linear transcripts that are relevant for HCC and beyond. As a final note, the authors should also be commended that their long list of circular RNAs compiled through their extensive RNAseq analysis confirmed the existence of some of the earliest circular RNAs identified by RT/PCR more than 20 years ago (13).
Acknowledgements
Funding: The author’s laboratory is funded by the Swedish Cancer Society and the Swedish Childhood Cancer Foundation.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Lei Liu (Department of Liver Disease and Digestive Interventional Radiology, Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2018.07.08). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Cocquerelle C, Daubersies P, Majerus MA, et al. Splicing with inverted order of exons occurs proximal to large introns. Embo J 1992;11:1095-8. [PubMed]
- Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 1996;93:6536-41. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Piwecka M, Glazar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017;357. [PubMed]
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 2018;68:1214-27. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Su X, Wang H, Ge W, et al. An In Vivo Method to Identify microRNA Targets Not Predicted by Computation Algorithms: p21 Targeting by miR-92a in Cancer. Cancer Res 2015;75:2875-85. [Crossref] [PubMed]
- Wang B, Hsu SH, Majumder S, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 2010;29:1787-97. [Crossref] [PubMed]
- Wang Y, Qin J, Liu Q, et al. SNF2H promotes hepatocellular carcinoma proliferation by activating the Wnt/beta-catenin signaling pathway. Oncol Lett 2016;12:1329-36. [Crossref] [PubMed]
- Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 1997;17:2985-93. [Crossref] [PubMed]
Cite this article as: Zaphiropoulos PG. Circles in action, circles in function. AME Med J 2018;3:81.

