
Personalised treatment for prostate cancer patients: are we there yet?
Prostate cancer (PCa) is an extremely heterogeneous disease—a small proportion of patients present with rapidly progressing, aggressive disease, referred to as unfavourable-risk PCa; whilst more commonly the disease progresses far more slowly and does not present an immediate health risk (1). This latter category, known as favourable-risk PCa, is not typically treated through intervention—instead, the recommendation for favourable-risk patients is monitoring via active surveillance (AS) (2). Generally, AS strategies are effective (3), although long-term surveillance studies have demonstrated PCa progression, in the form of disease grade reclassification (GR), in around 25% of cases by 10 years post-diagnosis (4,5). This GR risk is increased further in those of African-American race or advanced age (5,6). This propensity for PCa to progress during AS in some patients but not others, and particularly the association of GR with a particular ethnic group, suggests that underlying genetic factors may be important in disease progression, even in favourable-risk cases. Interrogating this heterogeneity provides a promising route to further stratify patients by their risk of disease progression, to better inform clinical assessment and lead to personalised treatment options.
ATM (ataxia-telangiectasia mutated), BRCA1 (breast cancer 1) and BRCA2 (breast cancer 2) are core components of the homology-directed repair pathway, one component of the DNA damage response—a simplified depiction is provided in Figure 1. ATM, a serine/threonine kinase, detects DNA damage and co-ordinates the cellular response by enabling numerous key damage-response pathways such as apoptosis and homology-directed repair (Figure 1A) (7-9). BRCA1 activates processes of homologous repair by recruiting effectors such as MRN and PALB2/BRCA2 (Figure 1B) (8,10). BRCA2 is a DNA-binding protein which contains a number of RAD51-binding repeats, allowing RAD51 localisation and filament formation for strand invasion (Figure 1C) (8,11).
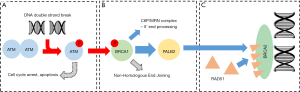
As one may predict given their importance in genome stability, ATM, BRCA1 and BRCA2 are known tumour suppressor genes, and their involvement with PCa is well supported (12-14). Particularly notable when considering sources of PCa heterogeneity, germline mutations in this three-gene panel have been specifically linked to aggressive and castration-resistance PCa cases and most recently by Na et al. (14), who demonstrated that germline mutational status was predictive not only of disease aggression, but also age of PCa-specific death and time to death after diagnosis.
Utilising this three gene panel, Carter et al. (4) set out to investigate whether it was possible to predict progression in favourable-risk PCa patients during AS. A cohort of 1,211 patients total was assembled, from the Johns Hopkins and North Shore Health Systems. Over the course of the study, on average 4 years at follow-up, GR was observed in 289 (23.9%) patients. BRCA2 mutation was significantly (P=0.03) associated with GR in six of eleven carriers. Neither ATM (P>0.99) not BRCA1 (P=0.3) mutational status alone significantly associated with GR. When analysing the data in terms of a three gene panel, mutation in any one gene was associated with GR occurrence (P=0.04). Relative risk estimates calculated that patients carrying a germline mutation in either ATM, BRCA or BRCA2 are approximately twice as likely to undergo GR during AS.
The conclusion drawn by Carter et al. is that mutations in the three gene panel are predictive of GR in patients during AS, and indeed this is supported by the data presented. That said, an alternative interpretation is that BRCA2 germline mutations alone are associated with GR, as the other members of the panel do not appear to be more frequently mutated in reclassified patients. It is important to point out that one limitation of the study, as noted by the authors, is the small number of carriers of mutations—only 26 patients carried a germline mutation in any of the selected genes. It is possible that these low numbers influenced results, as only five and ten patients carried mutations in ATM and BRCA1 respectively. To build on the groundwork laid by this study for understanding GR in patients, it will be important to expand the patient base considered to confirm specific mutational backgrounds facilitating progression.
Interestingly, BRCA2 mutation was also significantly (P=0.01) associated specifically with severe changes in grade group (from group 1 to group 3 or higher). Indeed, patients carrying a BRCA2 mutation were at a five-fold greater risk of severe GR versus non-carriers. In comparison, severe GR was only 2.5 times more likely in patients carrying a germline mutation in the three-gene panel versus the general population. As such, one could conclude that, while the three gene panel is associated with GR during AS, BRCA2 in particular is associated with larger scale GR. This is in agreement with current thinking on the role of BRCA2 in PCa, as studies have previously shown a link between BRCA2 mutation, earlier age of PCa diagnosis (15), and rapid disease progression (16,17).
Given the evidence presented both by Carter et al. and in other recent studies (14), there is a firm rationale for screening PCa patients before advising AS, especially those from at-risk populations with a family history of known or suspected BRCA2 mutation. Not only would screening patients allow for better stratification to predict clinical outcomes, but it opens a clear window for the application of personalised medicine. In particular, the loss of DNA repair capacity creates the possibility of creating synthetic lethality in rapidly dividing cancer cells by inducing DNA damage. Traditionally, platinum-based compounds have been used to this effect—platinum forms adducts with DNA, distorting the double helix and triggering damage response (18). More recently, PARP inhibitors, which cause double strand break accumulation, have emerged as an alternative. Indeed, PCa patients carrying mutations in DNA damage genes have been shown to respond more favourably to treatment with Olaparib (a PARP inhibitor) in a phase 2 clinical trial (19). In particular, all patients who carried a BRCA2 mutation responded well to treatment. Further, Olaparib has been FDA approved for treatment of ovarian and breast cancer patients with germline BRCA mutations (20). Clearly, induction of DNA damage represents a viable strategy for precision targeting of cancers in which DNA repair genes are mutated.
A secondary objective of Carter et al. was to investigate germline mutations in 51 other DNA repair genes, of which none were found to significantly associate with GR. Once more however, this result may have been impacted by low number of carriers investigated. Potentially interesting genes such as MSH2 and MSH6, both vital in DNA mismatch repair and reportedly present in 12% of patient tumours, have no germline mutations represented in the Carter et al. patient population (20). Another promising candidate progression-related gene is XPC, which is part of the nucleotide excision repair pathway, and variants of which are related to PCa risk (21). Here, XPC is only mutated in two patients, but both presented GR. It is fair to say, then, that DNA repair pathways outside of homology-directed repair have not been conclusively proven not to associate with progression in AS patients. As such, while screening for the three gene panel proposed here may provide clinical utility, it would be a worthwhile endeavour to re-investigate alternate DNA repair pathways in a larger cohort.
Carter et al. take some notable steps towards addressing the presently unanswered question of why PCa presents so differently in different patients. Indeed, the authors provide compelling evidence of an association between their three-gene panel, in particular BRCA2, with GR during surveillance. This has potentially important clinical impact, as it adds weight to the growing suggestion that screening for underlying DNA repair mutations could assist in making informed therapeutic decisions to best address the needs of each specific patient. So, while we may not have arrived yet at a personalised approach for diagnosing and treating PCa patients, the study by Carter et al. and related recent studies on mutations in DNA repair genes shows a clear direction of travel going forward.
Acknowledgments
Funding: Work in the McEwan laboratory is supported by local charities Friends of Anchor, Cranes and NHS Grampian.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xiao Li (Department of Urology, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & Affiliated Cancer Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2018.12.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sanda MG, Cadeddu JA, Kirkby E, et al. Clinically localized prostate cancer: AUA/ASTRO/SUO guideline. Part I: risk stratification, shared decision making, and care options. J Urol 2018;199:683-90. [Crossref] [PubMed]
- Chen RC, Rumble RB, Jain S. Active surveillance for the management of localized prostate cancer (Cancer Care Ontario guideline): American Society of Clinical Oncology clinical practice guideline endorsement summary. J Oncol Pract 2016;12:267-9. [Crossref] [PubMed]
- Klotz L, Vesprini D, Sethukavalan P, et al. Long-term follow-up of a large active surveillance cohort of patients with prostate cancer. J Clin Oncol 2015;33:272-7. [Crossref] [PubMed]
- Carter HB, Helfand B, Mamawala M, et al. Germline Mutations in ATM and BRCA1/2 Are Associated with Grade Reclassification in Men on Active Surveillance for Prostate Cancer. Eur Urol 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Tosoian JJ, Mamawala M, Epstein JI, et al. Intermediate and Longer-Term Outcomes From a Prospective Active-Surveillance Program for Favorable-Risk Prostate Cancer. J Clin Oncol 2015;33:3379-85. [Crossref] [PubMed]
- Sundi D, Faisal FA, Trock BJ, et al. Reclassification rates are higher among African American men than Caucasians on active surveillance. Urology 2015;85:155-60. [Crossref] [PubMed]
- Lavin MF, Birrell G, Chen P, et al. ATM signaling and genomic stability in response to DNA damage. Mutat Res 2005;569:123-32. [Crossref] [PubMed]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 2011;12:68-78. [Crossref] [PubMed]
- Barlow C, Brown KD, Deng C, et al. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat Genet 1997;17:453. [Crossref] [PubMed]
- Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 2006;34:1416-26. [Crossref] [PubMed]
- Wong AK, Pero R, Ormonde PA, et al. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem 1997;272:31941-4. [Crossref] [PubMed]
- Robinson D, Van Allen EM, Wu Y, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215-28. [Crossref] [PubMed]
- Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 2016;375:443-53. [Crossref] [PubMed]
- Na R, Zheng SL, Han M, et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol 2017;71:740-7. [Crossref] [PubMed]
- Edwards SM, Kote-Jarai Z, Meitz J, et al. Two percent of men with early-onset prostate cancer harbor germline mutations in the BRCA2 gene. Am J Hum Genet 2003;72:1-12. [Crossref] [PubMed]
- Mitra A, Fisher C, Foster C, et al. Prostate cancer in male BRCA1 and BRCA2 mutation carriers has a more aggressive phenotype. Br J Cancer 2008;98:502-7. [Crossref] [PubMed]
- Narod SA, Neuhausen S, Vichodez G, et al. Rapid progression of prostate cancer in men with a BRCA2 mutation. Br J Cancer 2008;99:371-4. [Crossref] [PubMed]
- Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 2005;4:307-20. [Crossref] [PubMed]
- Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373:1697-708. [Crossref] [PubMed]
- Pritchard CC, Morrissey C, Kumar A, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun 2014;5:4988. [Crossref] [PubMed]
- Hirata H, Hinoda Y, Tanaka Y, et al. Polymorphisms of DNA repair genes are risk factors for prostate cancer. Eur J Cancer 2007;43:231-7. [Crossref] [PubMed]
Cite this article as: Gillen AD, McEwan IJ. Personalised treatment for prostate cancer patients: are we there yet? AME Med J 2019;4:2.

