Association of STAT3 polymorphism with tyrosine kinase inhibitors-induced safety and efficacy in patients with metastatic renal cell carcinoma: a systematic review
Introduction
Renal cell carcinoma (RCC) represents one of the most prevalent malignancies worldwide with its incidence continue to rise annually (1). Clear renal cell carcinoma (cRCC) constitutes approximately 83% of all RCC cases, which lead to the second mortality among patients suffering from urologic neoplasms (2,3). Although RCC can be clearly diagnosed at its early stage and cured by partial nephrectomy, approximately 30% and 20–40% patients still develop metastasis and recurrence after resection (4,5).
Because of its insensitivity to conventional chemotherapy and radiotherapy, molecular-targeted drugs have emerged as a predominant therapy for patients with metastatic renal cell carcinoma (mRCC) (6-9). Among various targeting-agents subtypes, the use of multiple tyrosine kinase inhibitors (mTKIs) is the standard first-line therapeutic option, including sorafenib, sunitinib, axitinib and so forth (10-13). Although certain mTKIs have been applied to clinical therapy, several problems such as the risk of adverse events and efficacy-loss have emerged during the current clinical trials (14-16). In a clinical study consisting of 104 mRCC patients with first-line TKIs treatment, 71.7% of patients had one or more toxicities such as arterial hypertension, hypothyroidism and hand-foot syndrome, only 18.3% suffered from any of the selected toxicities (17). A randomized phase 3 trial assigned 615 patients with mRCC and indicated that Grade 3 or 4 adverse events were more frequent in the sunitinib group (60.5%) than in the placebo group (19.4%) (18).
Signal transducer and activator of transcription (STAT) 3, as a member of STAT family, was reported to play a pivotal role in signal transduction as well as transcription activity (19). STAT3 has also been verified to involve in regulating cellular differentiation, proliferation, and survival (20). Furthermore, aberrant dysfunction of STAT3 is often involved in genitourinary tumors, including prostate cancer, bladder cancer, and kidney cancer (21). In RCC, STAT3 can participate in regulating tumor growth, invasion, and promoting tumor angiogenesis. For instance, Masuda et al quantified STAT3 mRNA levels in RCC patients and found that STAT3 was downregulated in tumor tissues than normal tissues (22). Zhou et al. analyzed the gene expression profiles of RCC and found activation of STAT3 promoted tumor growth via ERK signaling pathways (23).
In addition, as a point of convergence for the signaling pathways downstream of numerous tyrosine kinases, STAT3 polymorphism has been associated with treatment response to TKIs in mRCC, as well as the incidence of adverse events (9,24). In a retrospective study of Japanese, STAT3 polymorphism was related to sunitinib-induced stomatitis in patients with mRCC (25). Moreover, study of Yamamoto et al suggested that STAT3 polymorphism could predict hand-foot skin reactions (HFSR) and tumor response to TKIs treatment in mRCC patients (9,26). However, the function of STAT3 polymorphism involved in TKIs-treated safety and efficacy is still controversial. Therefore, we conducted this systematic review to obtain a better understanding of the correlation.
Methods
Search strategy
We searched several electronic databases including PubMed, EMBASE and Web of Science to identify relevant literature up date to April 30th, 2017. A “PRISMA” guideline was applied to the literature screening process in this systematic review (27).
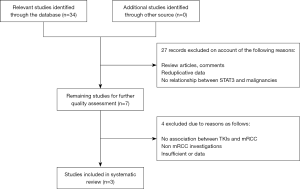
Primarily, only studies published in English could be included to in this systematic review. For the literature retrieval, following medical subject and text words were utilized: (“STAT3”, or “signal transducer and activator of transcription 3”), (“genetic polymorphism”, or “SNP”, or “single nucleotide polymorphism”), and (“renal cell carcinoam”, or “metastatic renal cell carcinoma”, or “mRCC”, or “RCC”). To minimize article omissions, the reference lists of eligible studies were manually screened for additional publications. A flow diagram of the study selection process is presented in Figure 1.
Quality assessment
We used a critical review checklist to systematically assess the quality of all studies included. The following inclusion criteria should be contained: (I) the relationship between STAT3 polymorphism and TKIs-induced events in mRCC; (II) clear STAT3 polymorphism sites in eligible articles; (III) non-overlapping available data in different studies. Conversely, studies were excluded when they did not cover the points above.
Data extraction
All relevant data of included studies were identified by two investigators (C Miao and A Xu) independently to rule out any discrepancy. Extracted data were reviewed by a third investigator (Y Zheng). The following elements were recorded: (I) the first author’s name, publication year; (II) ethnicity; (III) patient numbers (case and control); (IV) treatment of TKIs; (V) STAT3 polymorphism sites; (VI); TKIs-induced events; odds ratios (ORs) with 95% confidence interval (CI) between STAT3 polymorphism and TKIs-induced events were extracted from enrolled studies. All data information mentioned above was comprehensively presented in Tables 1,2.
Table 1
| First author | Year | Age | Gender | Ethnicity | Patients number | Treatment | Detected sample | STAT3 polymorphism | Events | MSKCC risk group | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Total | Control | Case | Favorable | Intermediate | Poor | ||||||||
| Watanabe (25) | 2017 | 67.5 [41–89] | 36 | 16 | Asian | 52 | 30 | 22 | Sunitinib | Blood | rs744166/rs4796793 | Stomatitis | NR | NR | NR |
| Yamamoto (9) | 2016 | 70.6 [40–90] | 43 | 17 | Asian | 60 | 14 | 46 | Sunitinib/sorafenib/axitinib | Blood | rs4796793 | HFSR | 18 | 39 | 3 |
| Yamamoto (26) | 2016 | 70.9 [40–90] | 34 | 16 | Asian | 50 | 33 | 17 | Sunitinib/sorafenib/axitinib | Blood | rs744166/rs4796793/rs9891119 | Response | 25 | 24 | 1 |
HFSR, hand-foot skin reactions; MSKCC, Memorial Sloan Kettering Cancer Center; NR, not reported.
Table 2
| First author | Year | Events | STAT3 polymorphism | Allele/genotype | Non-events | Events | OR | 95% CI | P value |
|---|---|---|---|---|---|---|---|---|---|
| Safety | |||||||||
| Watanabe (25) | 2017 | Stomatitis | rs744166 | T | 30 | 31 | 2.39 | 1.05–5.43 | 0.045* |
| C | 39 | 13 | |||||||
| TT | 10 | 11 | 2 | 0.65–6.19 | 0.26 | ||||
| TC + CC | 20 | 11 | |||||||
| TT + TC | 20 | 20 | 5 | 0.97–25.8 | 0.051 | ||||
| CC | 10 | 2 | |||||||
| rs4796793 | C | 31 | 31 | 2.23 | 0.98–5.08 | 0.069 | |||
| G | 29 | 13 | |||||||
| CC | 10 | 11 | 2 | 0.65–6.19 | 0.26 | ||||
| CG + GG | 20 | 11 | |||||||
| CC + CG | 21 | 20 | 4.29 | 0.82–22.3 | 0.092 | ||||
| GG | 9 | 2 | |||||||
| Yamamoto (9) | 2016 | HFSR | rs4796793 | G | 18 | 27 | 4.33 | 1.80–10.45 | 0.001* |
| C | 10 | 65 | |||||||
| GG | 6 | 3 | 10.75 | 2.38–48.07 | 0.001* | ||||
| CC + CG | 8 | 43 | |||||||
| CG + GG | 12 | 24 | 5.5 | 1.21–24.16 | 0.025* | ||||
| CC | 2 | 22 | |||||||
| HFSR severity | rs4796793 | CG + GG | 14 | 0 | 0.8 | 0.25–2.58 | 0.769 | ||
| CC | 14 | 18 | |||||||
| HFSR with sunitinib | rs4796793 | GG | 5 | 2 | 15 | 2.37–93.28 | 0.002* | ||
| GC + CC | 4 | 24 | |||||||
| CG + GG | 7 | 12 | 4.08 | 0.78–20.45 | 0.101 | ||||
| CC | 2 | 14 | |||||||
| Efficacy | |||||||||
| Yamamoto (26) | 2016 | Response | rs4796793 | G | 33 | 8 | 3.25 | 1.30–8.07 | 0.018* |
| C | 33 | 26 | |||||||
| GG | 8 | 1 | 5.12 | 0.73–33.84 | 0.141 | ||||
| CC + CG | 25 | 16 | |||||||
| CC | 8 | 10 | 4.46 | 1.31–15.28 | 0.028* | ||||
| CG + GG | 25 | 7 | |||||||
| rs744166 | C | 33 | 9 | 2.78 | 1.14–6.74 | 0.032* | |||
| T | 33 | 25 | |||||||
| CC | 8 | 2 | 2.4 | 0.50–11.20 | 0.461 | ||||
| TT + TC | 25 | 15 | |||||||
| TT | 8 | 10 | 4.46 | 1.31–15.28 | 0.028* | ||||
| TC + CC | 25 | 7 | |||||||
| rs9891119 | C | 33 | 9 | 2.78 | 1.14–6.74 | 0.032* | |||
| A | 33 | 25 | |||||||
| CC | 8 | 2 | 2.4 | 0.50–11.20 | 0.461 | ||||
| CA + AA | 25 | 15 | |||||||
| AA | 8 | 10 | 4.46 | 1.31–15.28 | 0.028* | ||||
| CC + CA | 25 | 7 |
*, P<0.05. TKI, tyrosine kinase inhibitor; mRCC, metastatic renal cell carcinoma; HFSR, hand-foot skin reactions; OR, odds ratio; CI, confidence interval.
Results
Summary of eligible studies
A total of three relevant studies were finally applicable to this systematic review, including 162 patients (113 males and 49 females). The mean age was 69.7, ranging from 40 to 90. All included studies were performed in Asian population. DNA was isolated from blood samples of all patients except one from blood mononuclear cells. Furthermore, risk group stratification was reported on basis of Memorial Sloan Kettering Cancer Center (MSKCC) prognostic criteria in two studies (28). The detailed characteristics of enrolled studies were summarized in Table 1.
The present series comprised RCC patients who were diagnosed with metastases foci and subsequently treated with TKIs as a first line therapy. Among the three studies, patients of 2 studies received multiple TKIs treatment including sunitinib, sorafenib, and axitinib, and 1 reported only sunitinib as implemented. Besides, the TKIs-induced events of three enrolled studies consisted of stomatitis, responder, and HFSR, respectively. Evaluation of these adverse events was generally determined using the National Cancer Institute Common Terminology Criteria for Adverse Events, version 4.0 or Response Evaluation Criteria in Solid Tumors ver.1.1, basing on patients' medical records. Among the three studies, several STAT3 SNP sites including rs744166, rs4796793 and rs9891119 were reported to correlate with events caused by TKIs treatments. Association data between the two were exhibited in Table 2.
Association between STAT3 polymorphism and TKIs-induced safety
Our systematic review suggested that STAT3 polymorphism served as a predictive factor for several adverse events induced by first line TKIs therapy in patients with mRCC. Watanabe et al. (25) conducted a retrospective analysis and reported the association between sunitinib-induced stomatitis and STAT3 polymorphism. STAT3 rs744166 (allele T vs. C) had a tendency to participate in sunitinib-induced stomatitis development (OR =2.39; 95% CI, 1.05–5.43; P=0.045; Table 2). However, no significant association was observed between rs744166 (genotype TT vs. TC + CC, TT + TC vs. CC), rs4796793 (allele C vs. G; genotype CC vs. CG + GG, CC + CG vs. GG; Table 2) and stomatitis risk.
Furthermore, Yamamoto et al. (9) reported that STAT3 rs4796793 genotype appeared to be a novel factor for TKIs-induced HFSR in patients with mRCC. Among their results, rs4796793 G allele was related to deferred HFSR development (OR =4.33; 95% CI, 1.80–10.45; P=0.001; Table 2). rs4796793 (genotype GG vs. CC + CG) exerted a negative association with HFSR (OR =10.75; 95% CI, 2.38–48.07; P=0.001), as well as CG + GG vs. CC (OR =5.5; 95% CI, 1.21–24.16; P=0.025; Table 2). Moreover, in patients treated with sunitinib as initial therapy, rs4796793 G allele homozygosity revealed a significant adverse relation (OR =15.00; 95% CI, 2.37–93.28, P=0.002; Table 2) with the development of HFSR. Nevertheless, similar relevant conclusion was not obtained between all rs4796793 models and HFSR severity in mTKIs-treated mRCC patients.
Association between STAT3 polymorphism and TKIs-induced efficacy
In another study of Yamamoto et al. (26), they declared that STAT3 polymorphisms were associated with treatment response to TKIs. STAT3 rs4796793 C allele was significantly correlated with the treatment response (OR =3.25; 95% CI, 1.30–8.07; P=0.018; Table 2). Such connection was also observed between rs744166 (allele C vs. T) and rs9891119 (allele C vs. A; Table 2). Moreover, the homozygosity of the C allele of rs4796793, the T allele of rs744166, and the A allele of rs989119 could also predict the mRCC response to TKIs therapy (Table 2).
Discussion
Nowadays, mTKIs have been widely used as the standard treatment for patients with mRCC, such as sorafenib, sunitinib, and axitinib (11,29,30). However, it is suggested that long-term treatment of mTKIs may result in severe adverse reactions, including hypertension, fatigue, and stomatitis, HFSR and other toxicities (9,25,31). Dysregulation of several proteins like STAT3, VEGF, and ABCG2 may account for the occurrence of these adverse events mediated through TKIs (25,32,33).
STAT3, which belongs to the STAT family, is involved in mediating cellular responses to cytokines (34). STAT3 activators comprise IL-6, EGF, S1P, Src family members and growth factor receptors that possess intrinsic tyrosine-kinase activity, such EGFRs, HGF receptor and PDGFR (35-38). Furthermore, STAT3 is activated by the phosphorylation of the tyrosine residue at position 705 by JAK (39). Activated pSTAT3 forms dimers and binds to specific DNA-response elements to promote the transcription of selected genes (21).
Recent investigations have revealed that STAT3 signaling pathways involves in the response to treatment in RCC. For instance, axitinib as one of the VEGFR-TKIs, has been reported to regulate antitumor immunity by down-regulating STAT3 expression (40). In addition, a RCC study in vitro has reported that the underlying mechanism of antitumor effects of sunitinib was partly attributed to the inhibition of STAT3 expression. In recent works, the association of STAT3 polymorphisms with various diseases as well as treatment toxicity has been confirmed in Asian population (41-44). In particular, STAT3 rs4796793 is known to modulate its mRNA expression in B lymphocyte cell lines as a consequence of its location in the 5’-flanking region of STAT3 (42). A previous study has also reported the incidence of mTKIs-induced skin toxicity by the inhibition of STAT3 activity in human epidermal keratinocytes in vitro (33). Therefore, different adverse events induced by mTKIs treatment in RCC patients may be partially ascribed to the STAT3 activity regulated by specific STAT3 polymorphisms.
In our study, we first reviewed the association between STAT3 polymorphisms and TKIs-induced safety and efficacy in patients with mRCC. Relevant events of available studies included the stomatitis, the development and severity of HFSR, and the response. Report of Watanabe indicated that only STAT3 rs744166 (allele T vs. C) are related to the development of stomatitis in sunitinib-treated mRCC patients. However, the probability of certain false positive in STAT3 rs744166 might exist in the positive results, because stomatitis development depended on various factors such as infections in immunocompromised patients, poor oral hygiene, nutritional deficiency, or steroid administration (45). Additionally, STAT3 polymorphisms were also associated with the development and severity of HFSR, which has emerged as a common toxicity in mRCC population treated with mTKIs. The progression of HFSR might be led by the low levels of STAT3 mRNA, as a result of decreased transcription of genes encoding various apoptosis suppressors, including survivin, myeloid cell leukemia 1 (Mcl-1), and B-cell lymphoma 2 (bcl-2), in keratinocytes (46-48). However, further confirmation of studies with large-scale sample size was required.
In consideration of its effects on TKIs-treated efficacy, three STAT3 polymorphisms (rs744166, rs4796793, rs9891119) were found to be associated with the treatment response to mTKIs in patients with mRCC. Differences in responses to mTKIs by different STAT3 polymorphisms might attribute to the regulation of T-cells apoptosis, based on the association between PDL-1 and STAT3 expression (49-51). Due to the limitation of study numbers, further high-quality investigations are needed to verify this conclusion.
Admittedly, our review still had several limitations for the following causes. First, only 3 eligible studies focusing on STAT3 polymorphism and TKIs-induced events were included in this systematic review. More clinical studies of high-quality and large sample size are required to strength our conclusion and make further confirmation. Second, we reported three different adverse events caused by TKIs in three individual investigations, which might lead to limited statistical validity and dispersed conclusion. Third, only Asian population was analyzed to evaluate the association. The deficiency of multiple ethnicities might weaken the credible power of the association. Further studies focusing on other ethnicities are also needed.
Acknowledgements
Funding: None.
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2017.05.09). Dr. Xiao Li serves as an unpaid Section Editor of AME Medical Journal. The other authors has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Capitanio U, Montorsi F. Renal cancer. Lancet 2016;387:894-906. [Crossref] [PubMed]
- Singer EA, Gupta GN, Srinivasan R. Update on targeted therapies for clear cell renal cell carcinoma. Curr Opin Oncol 2011;23:283. [Crossref] [PubMed]
- Zhuang J, Tu X, Cao K, et al. The expression and role of tyrosine kinase ETK/BMX in renal cell carcinoma. J Exp Clin Cancer Res 2014;33:25. [Crossref] [PubMed]
- Leibovich BC, Lohse CM, Crispen PL, et al. Histological subtype is an independent predictor of outcome for patients with renal cell carcinoma. J Urol 2010;183:1309-15. [Crossref] [PubMed]
- Pantuck AJ, Zisman A, Belldegrun AS. The changing natural history of renal cell carcinoma. J Urol 2001;166:1611-23. [Crossref] [PubMed]
- Godley P, Kim SW. Renal cell carcinoma. Curr Opin Oncol 2002;14:280-85. [Crossref] [PubMed]
- Blanco AI, Teh BS, Amato RJ. Role of radiation therapy in the management of renal cell cancer. Cancers 2011;3:4010-23. [Crossref] [PubMed]
- Hartmann JT, Bokemeyer C. Chemotherapy for renal cell carcinoma. AntiCancer Res 1999;19:1541-43. [PubMed]
- Yamamoto K, Shinomiya K, Ioroi T, et al. Association of Single Nucleotide Polymorphisms in STAT3 with Hand-Foot Skin Reactions in Patients with Metastatic Renal Cell Carcinoma Treated with Multiple Tyrosine Kinase Inhibitors: A Retrospective Analysis in Japanese Patients. Target Oncol 2016;11:93-9. [Crossref] [PubMed]
- Larkin J, Paine A, Tumur I, et al. Second-line treatments for the management of advanced renal cell carcinoma: systematic review and meta-analysis. Expert Opin Pharmacother 2013;14:27-39. [Crossref] [PubMed]
- Oya M, Tomita Y, Fukasawa S, et al. Overall survival of 1st‐line axitinib in metastatic renal cell carcinoma: Japanese subgroup analysis from phase II study. Cancer Sci 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Randrup Hansen C, Grimm D, Bauer J, et al. Effects and side effects of using sorafenib and sunitinib in the treatment of metastatic renal cell carcinoma. Int J Mol Sci 2017;18:461. [Crossref] [PubMed]
- Sonpavde G, Choueiri TK, Escudier B, et al. Sequencing of agents for metastatic renal cell carcinoma: can we customize therapy? Eur Urol 2012;61:307-16. [Crossref] [PubMed]
- Domagała-Haduch M, Cedrych I, Jasiówka M, et al. Analysis of adverse events of sunitinib in patients treated for advanced renal cell carcinoma. Arch Med Sci 2016;12:360-64. [Crossref] [PubMed]
- Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med 2007;356:125-34. [Crossref] [PubMed]
- Motzer RJ, Hutson TE, Tomczak P, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 2007;356:115-24. [Crossref] [PubMed]
- Iacovelli R, Verri E, Rocca MC, et al. Prognostic role of the cumulative toxicity in patients affected by metastatic renal cells carcinoma and treated with first-line tyrosine kinase inhibitors. Anticancer Drugs 2017;28:206-12. [Crossref] [PubMed]
- Ravaud A, Motzer RJ, Pandha HS, et al. Adjuvant sunitinib in high-risk renal-cell carcinoma after nephrectomy. N Engl J Med 2016;375:2246-54. [Crossref] [PubMed]
- Raz R, Durbin JE, Levy DE. Acute phase response factor and additional members of the interferon-stimulated gene factor 3 family integrate diverse signals from cytokines, interferons, and growth factors. J Biol Chem 1994;269:24391-95. [PubMed]
- Darnell JE. STATs and gene regulation. Science 1997;277:1630-35. [Crossref] [PubMed]
- Santoni M, Conti A, Piva F, et al. Role of STAT3 pathway in genitourinary tumors. Future Sci OA 2015;1:FSO15. [Crossref] [PubMed]
- Masuda A, Kamai T, Abe H, et al. Is Stat3 and/or p53 mRNA expression a prognostic marker for renal cell carcinoma? Biomed Res 2009;30:171-76. [Crossref] [PubMed]
- Zhou J, Deng Z, Chen Y, et al. Overexpression of FABP7 promotes cell growth and predicts poor prognosis of clear cell renal cell carcinoma. Urol Oncol 2015;33:113.e9-17. [Crossref] [PubMed]
- Xin H, Zhang C, Herrmann A, et al. Sunitinib inhibition of Stat3 induces renal cell carcinoma tumor cell apoptosis and reduces immunosuppressive cells. Cancer Res 2009;69:2506-13. [Crossref] [PubMed]
- Watanabe A, Yamamoto K, Ioroi T, et al. Association of Single Nucleotide Polymorphisms in STAT3, ABCB1, and ABCG2 with Stomatitis in Patients with Metastatic Renal Cell Carcinoma Treated with Sunitinib: A Retrospective Analysis in Japanese Patients. Biol Pharm Bull 2017;40:458-64. [Crossref] [PubMed]
- Yamamoto K, Ioroi T, Kanaya K, et al. STAT3 polymorphism rs4796793 may be a predictive factor of tumor response to multiple tyrosine kinase inhibitors in metastatic renal cell carcinoma in Japanese population. Med Oncol 2016;33:24-7. [Crossref] [PubMed]
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [Crossref] [PubMed]
- Motzer RJ, Bacik J, Murphy BA, et al. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 2002;20:289-96. [Crossref] [PubMed]
- Afriansyah A, Hamid AR, Mochtar CA, et al. Targeted Therapy for Metastatic Renal Cell Carcinoma. Acta Medica Indonesiana 2016;48:335-47. [PubMed]
- Roskoski R. Vascular endothelial growth factor (VEGF) and VEGF receptor inhibitors in the treatment of renal cell carcinomas. Pharmacol Res 2017;120:116-32. [Crossref] [PubMed]
- Di Lorenzo G, Porta C, Bellmunt J, et al. Toxicities of targeted therapy and their management in kidney cancer. Eur Urol 2011;59:526-40. [Crossref] [PubMed]
- Low S-K, Fukunaga K, Takahashi A, et al. Association study of a functional variant on ABCG2 gene with sunitinib-induced severe adverse drug reaction. PloS One 2016;11:e0148177. [Crossref] [PubMed]
- Yamamoto K, Mizumoto A, Nishimura K, et al. Association of toxicity of sorafenib and sunitinib for human keratinocytes with inhibition of signal transduction and activator of transcription 3 (STAT3). PloS One 2014;9:e102110. [Crossref] [PubMed]
- Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994;264:1415-21. [Crossref] [PubMed]
- Guo L, Chen C, Shi M, et al. Stat3-coordinated Lin-28–let-7–HMGA2 and miR-200–ZEB1 circuits initiate and maintain oncostatin M-driven epithelial–mesenchymal transition. Oncogene 2013;32:5272-82. [Crossref] [PubMed]
- Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Statl and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995;82:241-50. [Crossref] [PubMed]
- West NR, Murray JI, Watson PH. Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene 2014;33:1485-94. [Crossref] [PubMed]
- Zhong Z, Wen Z, Darnell JE Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994;264:95-8. [Crossref] [PubMed]
- Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene 2004;23:8017-23. [Crossref] [PubMed]
- Yuan H, Cai P, Li Q, et al. Axitinib augments antitumor activity in renal cell carcinoma via STAT3-dependent reversal of myeloid-derived suppressor cell accumulation. Biomed Pharmacother 2014;68:751-56. [Crossref] [PubMed]
- Eto M, Kamba T, Miyake H, et al. STAT3 polymorphism can predict the response to interferon-α therapy in patients with metastatic renal cell carcinoma. Eur Urol 2013;63:745-52. [Crossref] [PubMed]
- Ito N, Eto M, Nakamura E, et al. STAT3 polymorphism predicts interferon-alfa response in patients with metastatic renal cell carcinoma. J Clin Oncol 2007;25:2785-91. [Crossref] [PubMed]
- Sato K, Shiota M, Fukuda S, et al. Strong evidence of a combination polymorphism of the tyrosine kinase 2 gene and the signal transducer and activator of transcription 3 gene as a DNA-based biomarker for susceptibility to Crohn’s disease in the Japanese population. J Clin Immunol 2009;29:815-25. [Crossref] [PubMed]
- Yamazaki K, Umeno J, Takahashi A, et al. A genome-wide association study identifies 2 susceptibility loci for Crohn's disease in a Japanese population. Gastroenterology 2013;144:781-88. [Crossref] [PubMed]
- Kollmannsberger C, Bjarnason G, Burnett P, et al. Sunitinib in metastatic renal cell carcinoma: recommendations for management of noncardiovascular toxicities. Oncologist 2011;16:543-53. [Crossref] [PubMed]
- Aoki Y, Feldman GM, Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood 2003;101:1535-42. [Crossref] [PubMed]
- Epling-Burnette PK, Liu JH, Catlett-Falcone R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest 2001;107:351-62. [Crossref] [PubMed]
- Stephanou A, Brar BK, Knight RA, et al. Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters. Cell Death Differ 2000;7:329-30. [Crossref] [PubMed]
- Marzec M, Zhang Q, Goradia A, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad Sci U S A 2008;105:20852-57. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Parsa AT, Waldron JS, Panner A, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med 2007;13:84-88. [Crossref] [PubMed]
Cite this article as: Miao C, Xu A, Zheng Y, Liang C, Zhu J, Tian Y, Ma B, Lu P, Xu W, Li X. Association of STAT3 polymorphism with tyrosine kinase inhibitors-induced safety and efficacy in patients with metastatic renal cell carcinoma: a systematic review. AME Med J 2017;2:69.